1A96
 
 | | XPRTASE FROM E. COLI WITH BOUND CPRPP AND XANTHINE | | Descriptor: | 1-ALPHA-PYROPHOSPHORYL-2-ALPHA,3-ALPHA-DIHYDROXY-4-BETA-CYCLOPENTANE-METHANOL-5-PHOSPHATE, BORIC ACID, MAGNESIUM ION, ... | | Authors: | Vos, S, Parry, R.J, Burns, M.R, De Jersey, J, Martin, J.L. | | Deposit date: | 1998-04-16 | | Release date: | 1998-11-11 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structures of free and complexed forms of Escherichia coli xanthine-guanine phosphoribosyltransferase.
J.Mol.Biol., 282, 1998
|
|
3PUJ
 
 | | Crystal structure of the MUNC18-1 and SYNTAXIN4 N-Peptide complex | | Descriptor: | Syntaxin-4 N-terminal peptide, Syntaxin-binding protein 1 | | Authors: | Hu, S.-H, Christie, M.P, Saez, N.J, Latham, C.F, Jarrott, R, Lua, L.H.L, Collins, B.M, Martin, J.L. | | Deposit date: | 2010-12-05 | | Release date: | 2011-01-19 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.313 Å) | | Cite: | Possible roles for Munc18-1 domain 3a and Syntaxin1 N-peptide and C-terminal anchor in SNARE complex formation
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
4OCF
 
 | |
1BED
 
 | | STRUCTURE OF DISULFIDE OXIDOREDUCTASE | | Descriptor: | DSBA OXIDOREDUCTASE | | Authors: | Hu, S.-H, Martin, J.L. | | Deposit date: | 1996-09-16 | | Release date: | 1997-10-08 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of TcpG, the DsbA protein folding catalyst from Vibrio cholerae.
J.Mol.Biol., 268, 1997
|
|
4OCE
 
 | |
4OD7
 
 | |
1CJM
 
 | | HUMAN SULT1A3 WITH SULFATE BOUND | | Descriptor: | PROTEIN (ARYL SULFOTRANSFERASE), SULFATE ION | | Authors: | Bidwell, L.M, Mcmanus, M.E, Gaedigk, A, Kakuta, Y, Negishi, M, Pedersen, L, Martin, J.L. | | Deposit date: | 1999-04-18 | | Release date: | 1999-11-10 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of human catecholamine sulfotransferase.
J.Mol.Biol., 293, 1999
|
|
1G9S
 
 | | CRYSTAL STRUCTURE OF A COMPLEX BETWEEN E.COLI HPRT AND IMP | | Descriptor: | ANY 5'-MONOPHOSPHATE NUCLEOTIDE, HYPOXANTHINE PHOSPHORIBOSYLTRANSFERASE, INOSINIC ACID | | Authors: | Guddat, L.W, Vos, S, Martin, J.L, Keough, D.T, de Jersey, J. | | Deposit date: | 2000-11-27 | | Release date: | 2002-08-28 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structures of free, IMP-, and GMP-bound Escherichia coli hypoxanthine phosphoribosyltransferase.
Protein Sci., 11, 2002
|
|
1G9T
 
 | | CRYSTAL STRUCTURE OF E.COLI HPRT-GMP COMPLEX | | Descriptor: | ANY 5'-MONOPHOSPHATE NUCLEOTIDE, GUANOSINE-5'-MONOPHOSPHATE, HYPOXANTHINE PHOSPHORIBOSYLTRANSFERASE | | Authors: | Guddat, L.W, Vos, S, Martin, J.L, keough, D.T, de Jersey, J. | | Deposit date: | 2000-11-28 | | Release date: | 2002-08-28 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structures of free, IMP-, and GMP-bound Escherichia coli hypoxanthine phosphoribosyltransferase.
Protein Sci., 11, 2002
|
|
1PEN
 
 | | ALPHA-CONOTOXIN PNI1 | | Descriptor: | ALPHA-CONOTOXIN PNIA | | Authors: | Hu, S.-H, Gehrmann, J, Guddat, L.W, Alewood, P.F, Craik, D.J, Martin, J.L. | | Deposit date: | 1996-01-29 | | Release date: | 1997-04-21 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | The 1.1 A crystal structure of the neuronal acetylcholine receptor antagonist, alpha-conotoxin PnIA from Conus pennaceus.
Structure, 4, 1996
|
|
7RGV
 
 | |
2MBS
 
 | | NMR solution structure of oxidized KpDsbA | | Descriptor: | Thiol:disulfide interchange protein | | Authors: | Kurth, F, Rimmer, K, Premkumar, L, Mohanty, B, Duprez, W, Halili, M.A, Shouldice, S.R, Heras, B, Fairlie, D.P, Scanlon, M.J, Martin, J.L. | | Deposit date: | 2013-08-03 | | Release date: | 2013-12-11 | | Last modified: | 2024-11-27 | | Method: | SOLUTION NMR | | Cite: | Comparative Sequence, Structure and Redox Analyses of Klebsiella pneumoniae DsbA Show That Anti-Virulence Target DsbA Enzymes Fall into Distinct Classes.
Plos One, 8, 2013
|
|
3AX4
 
 | | Three-dimensional structure of lectin from Dioclea violacea and comparative vasorelaxant effects with Dioclea rostrata | | Descriptor: | 5-bromo-4-chloro-1H-indol-3-yl alpha-D-mannopyranoside, CALCIUM ION, MANGANESE (II) ION, ... | | Authors: | Bezerra, M.J.B, Bezerra, G.A, Martins, J.L, Nascimento, K.S, Nagano, C.S, Gruber, K, Assereuy, A.M, Delatorre, P, Rocha, B.A.M, Cavada, B.S. | | Deposit date: | 2011-03-29 | | Release date: | 2012-04-04 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.613 Å) | | Cite: | Crystal structure of Dioclea violacea lectin and a comparative study of vasorelaxant properties with Dioclea rostrata lectin
Int.J.Biochem.Cell Biol., 45, 2013
|
|
2W53
 
 | | Structure of SmeT, the repressor of the Stenotrophomonas maltophilia multidrug efflux pump SmeDEF. | | Descriptor: | REPRESSOR, SULFATE ION | | Authors: | Mate, M.J, Romero, A, Hernandez, A, Martinez, J.L. | | Deposit date: | 2008-12-04 | | Release date: | 2009-04-07 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural and Functional Analysis of Smet, the Repressor of the Stenotrophomonas Maltophilia Multidrug Efflux Pump Smedef.
J.Biol.Chem., 284, 2009
|
|
3P9T
 
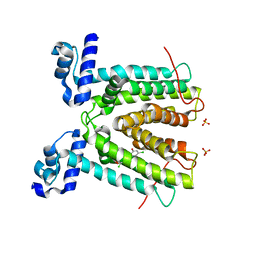 | | SmeT-Triclosan complex | | Descriptor: | Repressor, SULFATE ION, TRICLOSAN | | Authors: | Hernandez, A, Ruiz, F.M, Romero, A, Martinez, J.L. | | Deposit date: | 2010-10-18 | | Release date: | 2011-08-10 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | The Binding of Triclosan to SmeT, the Repressor of the Multidrug Efflux Pump SmeDEF, Induces Antibiotic Resistance in Stenotrophomonas maltophilia.
Plos Pathog., 7, 2011
|
|
5TLQ
 
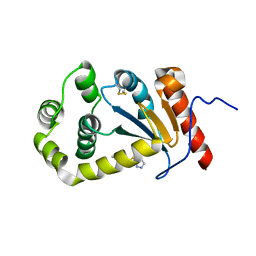 | | Model structure of the oxidized PaDsbA1 and 3-[(2-methylbenzyl)sulfanyl]-4H-1,2,4-triazol-4-amine complex | | Descriptor: | 3-[(2-methylbenzyl)sulfanyl]-4H-1,2,4-triazol-4-amine, Thiol:disulfide interchange protein DsbA | | Authors: | Mohanty, B, Rimmer, K.A, McMahon, R.M, Headey, S.J, Vazirani, M, Shouldice, S.R, Coincon, M, Tay, S, Morton, C.J, Simpson, J.S, Martin, J.L, Scanlon, M.S. | | Deposit date: | 2016-10-11 | | Release date: | 2017-04-12 | | Last modified: | 2024-11-06 | | Method: | SOLUTION NMR | | Cite: | Fragment library screening identifies hits that bind to the non-catalytic surface of Pseudomonas aeruginosa DsbA1.
PLoS ONE, 12, 2017
|
|
5U3E
 
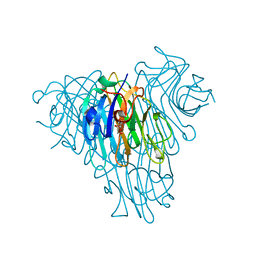 | | Crystal Structure of Native Lectin from Canavalia bonariensis Seeds (CaBo) complexed with alpha-methyl-D-mannoside | | Descriptor: | CALCIUM ION, Canavalia bonariensis seed lectin, MANGANESE (II) ION, ... | | Authors: | Silva, M.T.L, Osterne, V.J.S, Pinto-Junior, V.R, Santiago, M.Q, Araripe, D.A, Neco, A.H.B, Silva-Filho, J.C, Martins, J.L, Rocha, C.R.C, Leal, R.B, Nascimento, K.S, Cavada, B.S. | | Deposit date: | 2016-12-02 | | Release date: | 2017-08-23 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Canavalia bonariensis lectin: Molecular bases of glycoconjugates interaction and antiglioma potential.
Int. J. Biol. Macromol., 106, 2018
|
|
4WET
 
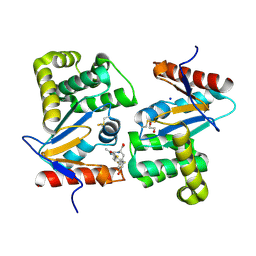 | | Crystal structure of E.Coli DsbA in complex with compound 16 | | Descriptor: | 1,2-ETHANEDIOL, N-({4-methyl-2-[4-(trifluoromethyl)phenyl]-1,3-thiazol-5-yl}carbonyl)-L-tyrosine, SODIUM ION, ... | | Authors: | Ilyichova, O.V, Scanlon, M.J. | | Deposit date: | 2014-09-11 | | Release date: | 2015-01-21 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Application of Fragment-Based Screening to the Design of Inhibitors of Escherichia coli DsbA.
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
6WI6
 
 | | Crystal structure of plantacyclin B21AG | | Descriptor: | MALONATE ION, Plantacyclin B21AG | | Authors: | Smith, A.T, Gor, M.C, Vezina, B, McMahon, R, King, G, Panjikar, S, Rehm, B, Martin, J. | | Deposit date: | 2020-04-08 | | Release date: | 2021-01-06 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure and site-directed mutagenesis of circular bacteriocin plantacyclin B21AG reveals cationic and aromatic residues important for antimicrobial activity.
Sci Rep, 10, 2020
|
|
3DYR
 
 | |
3E4H
 
 | | Crystal structure of the cyclotide varv F | | Descriptor: | Varv peptide F,Varv peptide F | | Authors: | Hu, S.H. | | Deposit date: | 2008-08-11 | | Release date: | 2009-02-10 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Combined X-ray and NMR Analysis of the Stability of the Cyclotide Cystine Knot Fold That Underpins Its Insecticidal Activity and Potential Use as a Drug Scaffold
J.Biol.Chem., 284, 2009
|
|
3OQ7
 
 | | Crystal Structures of Multidrug-Resistant Clinical Isolate 769 HIV-1 Protease Variants | | Descriptor: | HIV-1 Protease | | Authors: | Yedidi, R.S, Proteasa, G, Martinez-Cajas, J.L, Vickrey, J.F, Martin, P.D, Wawrzak, Z, Kovari, L.C. | | Deposit date: | 2010-09-02 | | Release date: | 2011-04-06 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Contribution of the 80s loop of HIV-1 protease to the multidrug-resistance mechanism: crystallographic study of MDR769 HIV-1 protease variants.
Acta Crystallogr.,Sect.D, 67, 2011
|
|
3OQA
 
 | | Crystal Structures of Multidrug-Resistant Clinical Isolate 769 HIV-1 Protease Variants | | Descriptor: | HIV-1 Protease | | Authors: | Yedidi, R.S, Proteasa, G, Martinez-Cajas, J.L, Vickrey, J.F, Martin, P.D, Wawrzak, Z, Kovari, L.C. | | Deposit date: | 2010-09-02 | | Release date: | 2011-04-06 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Contribution of the 80s loop of HIV-1 protease to the multidrug-resistance mechanism: crystallographic study of MDR769 HIV-1 protease variants.
Acta Crystallogr.,Sect.D, 67, 2011
|
|
3PJ6
 
 | | Crystal Structures of Multidrug-Resistant Clinical Isolate 769 HIV-1 Protease Variants | | Descriptor: | HIV protease | | Authors: | Yedidi, R.S, Proteasa, G, Martinez-Cajas, J.L, Vickrey, J.F, Martin, P.D, Wawrzak, Z, Kovari, L.C. | | Deposit date: | 2010-11-08 | | Release date: | 2011-04-06 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Contribution of the 80s loop of HIV-1 protease to the multidrug-resistance mechanism: crystallographic study of MDR769 HIV-1 protease variants.
Acta Crystallogr.,Sect.D, 67, 2011
|
|
3OQD
 
 | | Crystal Structures of Multidrug-Resistant Clinical Isolate 769 HIV-1 Protease Variants | | Descriptor: | HIV-1 Protease | | Authors: | Yedidi, R.S, Proteasa, G, Martinez-Cajas, J.L, Vickrey, J.F, Martin, P.D, Wawrzak, Z, Kovari, L.C. | | Deposit date: | 2010-09-02 | | Release date: | 2011-04-06 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Contribution of the 80s loop of HIV-1 protease to the multidrug-resistance mechanism: crystallographic study of MDR769 HIV-1 protease variants.
Acta Crystallogr.,Sect.D, 67, 2011
|
|
