3E7J
 
 | | HeparinaseII H202A/Y257A double mutant complexed with a heparan sulfate tetrasaccharide substrate | | Descriptor: | 4-deoxy-alpha-L-threo-hex-4-enopyranuronic acid-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-alpha-D-glucopyranuronic acid-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, Heparinase II protein, ... | | Authors: | Shaya, D, Cygler, M. | | Deposit date: | 2008-08-18 | | Release date: | 2008-12-30 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Catalytic mechanism of heparinase II investigated by site-directed mutagenesis and the crystal structure with its substrate.
J.Biol.Chem., 285, 2010
|
|
7JQQ
 
 | | The bacteriophage Phi-29 viral genome packaging motor assembly | | Descriptor: | DNA (60-MER), DNA packaging protein, MAGNESIUM ION, ... | | Authors: | White, M.A, Woodson, M, Morais, M.C. | | Deposit date: | 2020-08-11 | | Release date: | 2021-05-19 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | A viral genome packaging motor transitions between cyclic and helical symmetry to translocate dsDNA.
Sci Adv, 7, 2021
|
|
8WBG
 
 | |
8WBH
 
 | |
8WBF
 
 | |
8WBC
 
 | |
8WBB
 
 | |
8WBE
 
 | |
8WBD
 
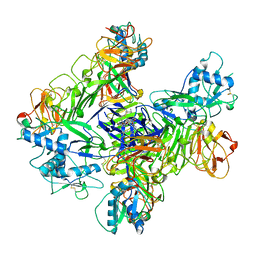 | |
6Z45
 
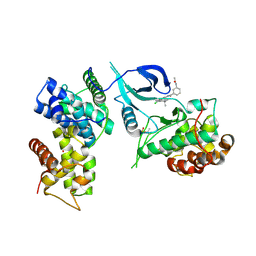 | | CDK9-Cyclin-T1 complex bound by compound 24 | | Descriptor: | (1~{S},3~{R})-3-acetamido-~{N}-[5-chloranyl-4-(5,5-dimethyl-4,6-dihydropyrrolo[1,2-b]pyrazol-3-yl)pyridin-2-yl]cyclohexane-1-carboxamide, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Cyclin-T1, ... | | Authors: | Ferguson, A, Collie, G.W. | | Deposit date: | 2020-05-22 | | Release date: | 2020-12-23 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (3.37 Å) | | Cite: | Discovery of AZD4573, a Potent and Selective Inhibitor of CDK9 That Enables Short Duration of Target Engagement for the Treatment of Hematological Malignancies.
J.Med.Chem., 63, 2020
|
|
1ZOA
 
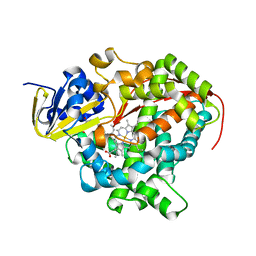 | | Crystal Structure Of A328V Mutant Of The Heme Domain Of P450Bm-3 With N-Palmitoylglycine | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Bifunctional P-450:NADPH-P450 reductase, GLYCEROL, ... | | Authors: | Hegda, A, Chen, B, Haines, D.C, Bondlela, M, Mullin, D, Graham, S.E, Tomchick, D.R, Machius, M, Peterson, J.A. | | Deposit date: | 2005-05-12 | | Release date: | 2006-08-01 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | A single active-site mutation of P450BM-3 dramatically enhances substrate binding and rate of product formation.
Biochemistry, 50, 2011
|
|
1ZO4
 
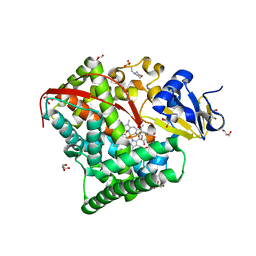 | | Crystal Structure Of A328S Mutant Of The Heme Domain Of P450BM-3 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Bifunctional P-450:NADPH-P450 reductase, GLYCEROL, ... | | Authors: | Hegda, A, Chen, B, Haines, D.C, Bondlela, M, Mullin, D, Graham, S.E, Tomchick, D.R, Machius, M, Peterson, J.A. | | Deposit date: | 2005-05-12 | | Release date: | 2006-08-01 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | A single active-site mutation of P450BM-3 dramatically enhances substrate binding and rate of product formation.
Biochemistry, 50, 2011
|
|
8JHF
 
 | | Native SUV420H1 bound to 167-bp nucleosome | | Descriptor: | DNA (160-MER), Histone H2A.Z, Histone H2B type 1-K, ... | | Authors: | Lin, F, Li, W. | | Deposit date: | 2023-05-23 | | Release date: | 2023-11-15 | | Last modified: | 2023-12-20 | | Method: | ELECTRON MICROSCOPY (3.68 Å) | | Cite: | Structural basis of nucleosomal H4K20 recognition and methylation by SUV420H1 methyltransferase.
Cell Discov, 9, 2023
|
|
8JHG
 
 | | Native SUV420H1 bound to 167-bp nucleosome | | Descriptor: | DNA (160-MER), Histone H2A type 1-B/E, Histone H2B type 1-K, ... | | Authors: | Lin, F, Li, W. | | Deposit date: | 2023-05-23 | | Release date: | 2023-11-22 | | Last modified: | 2023-12-20 | | Method: | ELECTRON MICROSCOPY (3.58 Å) | | Cite: | Structural basis of nucleosomal H4K20 recognition and methylation by SUV420H1 methyltransferase.
Cell Discov, 9, 2023
|
|
8FAZ
 
 | | Cryo-EM structure of the human BCDX2 complex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, DNA repair protein RAD51 homolog 2, DNA repair protein RAD51 homolog 3, ... | | Authors: | Jia, L, Wasmuth, E.V, Ruben, E.A, Sung, P, Rawal, Y, Greene, E.C, Meir, A, Olsen, S.K. | | Deposit date: | 2022-11-29 | | Release date: | 2023-06-21 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (2.3 Å) | | Cite: | Structural insights into BCDX2 complex function in homologous recombination.
Nature, 619, 2023
|
|
8I4V
 
 | | Cryo-EM structure of 5-subunit Smc5/6 arm region | | Descriptor: | DNA repair protein KRE29, E3 SUMO-protein ligase MMS21, Structural maintenance of chromosomes protein 5, ... | | Authors: | Qian, L, Jun, Z, Xiang, Z, Cheng, T, Zhaoning, W, Zhenguo, C, Wang, L. | | Deposit date: | 2023-01-21 | | Release date: | 2024-06-26 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (5.97 Å) | | Cite: | Cryo-EM structures of Smc5/6 in multiple states reveal its assembly and functional mechanisms.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8I13
 
 | | Cryo-EM structure of 6-subunit Smc5/6 | | Descriptor: | MMS21 isoform 1, NSE3 isoform 1, Non-structural maintenance of chromosomes element 1 homolog, ... | | Authors: | Qian, L, Jun, Z, Xiang, Z, Cheng, T, Zhaoning, W, Duo, J, Zhenguo, C, Wang, L. | | Deposit date: | 2023-01-12 | | Release date: | 2024-06-26 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (6.9 Å) | | Cite: | Cryo-EM structures of Smc5/6 in multiple states reveal its assembly and functional mechanisms.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8I4W
 
 | | Cryo-EM structure of 5-subunit Smc5/6 head region | | Descriptor: | DNA repair protein KRE29, Non-structural maintenance of chromosome element 5, Structural maintenance of chromosomes protein 5, ... | | Authors: | Qian, L, Jun, Z, Xiang, Z, Zhaoning, W, Cheng, T, Duo, J, Zhenguo, C, Wang, L. | | Deposit date: | 2023-01-21 | | Release date: | 2024-06-26 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (6.01 Å) | | Cite: | Cryo-EM structures of Smc5/6 in multiple states reveal its assembly and functional mechanisms.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8I4X
 
 | | Cryo-EM structure of 5-subunit Smc5/6 | | Descriptor: | E3 SUMO-protein ligase MMS21, Non-structural maintenance of chromosome element 5, Nse6, ... | | Authors: | Qian, L, Jun, Z, Xiang, Z, Zhaoning, W, Tong, C, Duo, J, Zhenguo, C, Wang, L. | | Deposit date: | 2023-01-21 | | Release date: | 2024-06-26 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (8.5 Å) | | Cite: | Cryo-EM structures of Smc5/6 in multiple states reveal its assembly and functional mechanisms.
Nat.Struct.Mol.Biol., 31, 2024
|
|
3Q3T
 
 | | Alkyl Amine Renin Inhibitors: Filling S1 from S3 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, ... | | Authors: | Wu, Z, McKeever, B. | | Deposit date: | 2010-12-22 | | Release date: | 2011-08-03 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Biphenyl/diphenyl ether renin inhibitors: Filling the S1 pocket of renin via the S3 pocket.
Bioorg.Med.Chem.Lett., 21, 2011
|
|
6IR8
 
 | | Rice WRKY/DNA complex | | Descriptor: | DNA (5'-D(P*GP*AP*TP*AP*TP*TP*TP*GP*AP*CP*CP*GP*GP*A)-3'), DNA (5'-D(P*TP*CP*CP*GP*GP*TP*CP*AP*AP*AP*TP*AP*TP*C)-3'), OsWRKY45, ... | | Authors: | Liu, J, Cheng, X, Wang, D. | | Deposit date: | 2018-11-12 | | Release date: | 2019-02-20 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis of dimerization and dual W-box DNA recognition by rice WRKY domain.
Nucleic Acids Res., 47, 2019
|
|
7W5Y
 
 | |
7W5X
 
 | | Cryo-EM structure of SoxS-dependent transcription activation complex with zwf promoter DNA | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | Lin, W, Feng, Y, Shi, J. | | Deposit date: | 2021-11-30 | | Release date: | 2022-10-26 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural basis of three different transcription activation strategies adopted by a single regulator SoxS.
Nucleic Acids Res., 50, 2022
|
|
7W5W
 
 | |
3Q4B
 
 | | Clinically Useful Alkyl Amine Renin Inhibitors | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, Renin, ... | | Authors: | Wu, Z, McKeever, B.M. | | Deposit date: | 2010-12-23 | | Release date: | 2011-11-30 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Discovery of VTP-27999, an Alkyl Amine Renin Inhibitor with Potential for Clinical Utility.
ACS Med Chem Lett, 2, 2011
|
|
