3PDU
 
 | |
3PEF
 
 | |
5JKJ
 
 | | Crystal structure of esterase E22 L374D mutant | | Descriptor: | Esterase E22 | | Authors: | Zhang, Y, Wang, P, Yao, Q. | | Deposit date: | 2016-04-26 | | Release date: | 2017-04-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structural basis for substrate recognition and catalysis of a novel esterase E22 with a homoserine transacetylase-like fold
To Be Published
|
|
2KHT
 
 | | NMR Structure of human alpha defensin HNP-1 | | Descriptor: | Neutrophil defensin 1 | | Authors: | Zhang, Y, Li, S, Doherty, T.F, Lubkowski, J, Lu, W, Li, J, Barinka, C, Hong, M. | | Deposit date: | 2009-04-11 | | Release date: | 2010-02-09 | | Last modified: | 2024-05-01 | | Method: | SOLID-STATE NMR | | Cite: | Resonance assignment and three-dimensional structure determination of a human alpha-defensin, HNP-1, by solid-state NMR.
J.Mol.Biol., 397, 2010
|
|
2KNQ
 
 | | Solution structure of E.Coli GspH | | Descriptor: | General secretion pathway protein H | | Authors: | Zhang, Y, Jin, C. | | Deposit date: | 2009-08-31 | | Release date: | 2010-09-08 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution structure of GspH
To be Published
|
|
5JKF
 
 | | Crystal structure of esterase E22 | | Descriptor: | Esterase E22 | | Authors: | Zhang, Y, Wang, P, Yao, Q. | | Deposit date: | 2016-04-26 | | Release date: | 2017-04-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.393 Å) | | Cite: | Structural basis for substrate recognition and catalysis of a novel esterase E22 with a homoserine transacetylase-like fold
To Be Published
|
|
3ZEE
 
 | | Electron cyro-microscopy helical reconstruction of Par-3 N terminal domain | | Descriptor: | PARTITIONING DEFECTIVE 3 HOMOLOG | | Authors: | Zhang, Y, Wang, W, Chen, J, Zhang, K, Gao, F, Gong, W, Zhang, M, Sun, F, Feng, W. | | Deposit date: | 2012-12-05 | | Release date: | 2013-10-16 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (6.1 Å) | | Cite: | Structural Insights Into the Intrinsic Self-Assembly of Par-3 N-Terminal Domain.
Structure, 21, 2013
|
|
2L3M
 
 | | Solution structure of the putative copper-ion-binding protein from Bacillus anthracis str. Ames | | Descriptor: | Copper-ion-binding protein | | Authors: | Zhang, Y, Winsor, J, Dubrovska, I, Anderson, W, Radhakrishnan, I, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2010-09-16 | | Release date: | 2011-01-12 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | To be published
To be Published
|
|
2LD5
 
 | |
2LLO
 
 | |
2L80
 
 | | Solution Structure of the Zinc Finger Domain of USP13 | | Descriptor: | Ubiquitin carboxyl-terminal hydrolase 13, ZINC ION | | Authors: | Zhang, Y, Zhou, C, Zhou, Z, Song, A, Hu, H. | | Deposit date: | 2010-12-28 | | Release date: | 2011-12-14 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Biochemical Characterization of the Ubiquitin Receptors in USP13 Reveals Different Catalytic Activation of Deubiquitination from Its Analogue USP5
To be Published
|
|
2LLQ
 
 | |
2LRJ
 
 | |
3LC8
 
 | |
2R86
 
 | | Crystal structure of PurP from Pyrococcus furiosus complexed with ATP | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, ADENOSINE-5'-TRIPHOSPHATE, PHOSPHATE ION, ... | | Authors: | Zhang, Y, White, R.H, Ealick, S.E. | | Deposit date: | 2007-09-10 | | Release date: | 2007-12-04 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure and function of 5-formaminoimidazole-4-carboxamide ribonucleotide synthetase from Methanocaldococcus jannaschii.
Biochemistry, 47, 2008
|
|
2R7K
 
 | | Crystal structure of FAICAR synthetase (PurP) from M. jannaschii complexed with AMPPCP and AICAR | | Descriptor: | 5-formaminoimidazole-4-carboxamide-1-(beta)-D-ribofuranosyl 5'-monophosphate synthetase, AMINOIMIDAZOLE 4-CARBOXAMIDE RIBONUCLEOTIDE, CHLORIDE ION, ... | | Authors: | Zhang, Y, White, R.H, Ealick, S.E. | | Deposit date: | 2007-09-09 | | Release date: | 2007-12-04 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure and function of 5-formaminoimidazole-4-carboxamide ribonucleotide synthetase from Methanocaldococcus jannaschii.
Biochemistry, 47, 2008
|
|
2R7M
 
 | | Crystal structure of FAICAR synthetase (PurP) from M. jannaschii complexed with AMP | | Descriptor: | 5-formaminoimidazole-4-carboxamide-1-(beta)-D-ribofuranosyl 5'-monophosphate synthetase, ADENOSINE MONOPHOSPHATE, CHLORIDE ION, ... | | Authors: | Zhang, Y, White, R.H, Ealick, S.E. | | Deposit date: | 2007-09-09 | | Release date: | 2007-12-04 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure and function of 5-formaminoimidazole-4-carboxamide ribonucleotide synthetase from Methanocaldococcus jannaschii.
Biochemistry, 47, 2008
|
|
2R84
 
 | | Crystal structure of PurP from Pyrococcus furiosus complexed with AMP and AICAR | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, ADENOSINE MONOPHOSPHATE, AMINOIMIDAZOLE 4-CARBOXAMIDE RIBONUCLEOTIDE, ... | | Authors: | Zhang, Y, White, R.H, Ealick, S.E. | | Deposit date: | 2007-09-10 | | Release date: | 2007-12-04 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure and function of 5-formaminoimidazole-4-carboxamide ribonucleotide synthetase from Methanocaldococcus jannaschii.
Biochemistry, 47, 2008
|
|
2R7L
 
 | | Crystal structure of FAICAR synthetase (PurP) from M. jannaschii complexed with ATP and AICAR | | Descriptor: | 5-formaminoimidazole-4-carboxamide-1-(beta)-D-ribofuranosyl 5'-monophosphate synthetase, ADENOSINE-5'-TRIPHOSPHATE, AMINOIMIDAZOLE 4-CARBOXAMIDE RIBONUCLEOTIDE, ... | | Authors: | Zhang, Y, White, R.H, Ealick, S.E. | | Deposit date: | 2007-09-09 | | Release date: | 2007-12-04 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure and function of 5-formaminoimidazole-4-carboxamide ribonucleotide synthetase from Methanocaldococcus jannaschii.
Biochemistry, 47, 2008
|
|
2R7N
 
 | | Crystal structure of FAICAR synthetase (PurP) from M. jannaschii complexed with ADP and FAICAR | | Descriptor: | 5-(formylamino)-1-(5-O-phosphono-beta-D-ribofuranosyl)-1H-imidazole-4-carboxamide, 5-formaminoimidazole-4-carboxamide-1-(beta)-D-ribofuranosyl 5'-monophosphate synthetase, ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Zhang, Y, White, R.H, Ealick, S.E. | | Deposit date: | 2007-09-09 | | Release date: | 2007-12-04 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure and function of 5-formaminoimidazole-4-carboxamide ribonucleotide synthetase from Methanocaldococcus jannaschii.
Biochemistry, 47, 2008
|
|
2R87
 
 | |
3LBS
 
 | |
6LN5
 
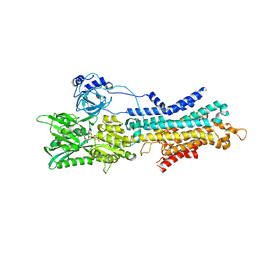 | | CryoEM structure of SERCA2b T1032stop in E1-2Ca2+-AMPPCP (class1) | | Descriptor: | CALCIUM ION, MAGNESIUM ION, PHOSPHOMETHYLPHOSPHONIC ACID ADENYLATE ESTER, ... | | Authors: | Zhang, Y, Tsutsumi, A, Watanabe, S, Inaba, K. | | Deposit date: | 2019-12-28 | | Release date: | 2020-08-26 | | Last modified: | 2020-09-16 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Cryo-EM structures of SERCA2b reveal the mechanism of regulation by the luminal extension tail.
Sci Adv, 6, 2020
|
|
6LN9
 
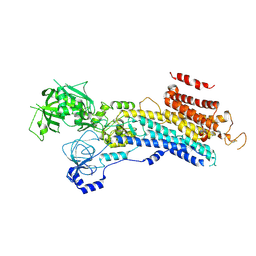 | | CryoEM structure of SERCA2b T1032stop in E2-BeF3- state (class2) | | Descriptor: | BERYLLIUM TRIFLUORIDE ION, MAGNESIUM ION, Sarcoplasmic/endoplasmic reticulum calcium ATPase 2 | | Authors: | Zhang, Y, Tsutsumi, A, Watanabe, S, Inaba, K. | | Deposit date: | 2019-12-28 | | Release date: | 2020-08-26 | | Last modified: | 2020-09-16 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Cryo-EM structures of SERCA2b reveal the mechanism of regulation by the luminal extension tail.
Sci Adv, 6, 2020
|
|
6LN7
 
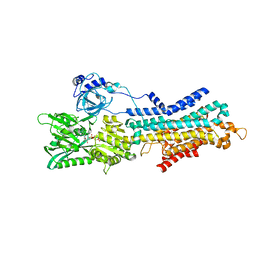 | | CryoEM structure of SERCA2b T1032stop in E1-2Ca2+-AMPPCP (class3) | | Descriptor: | CALCIUM ION, MAGNESIUM ION, PHOSPHOMETHYLPHOSPHONIC ACID ADENYLATE ESTER, ... | | Authors: | Zhang, Y, Tsutsumi, A, Watanabe, S, Inaba, K. | | Deposit date: | 2019-12-28 | | Release date: | 2020-08-26 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Cryo-EM structures of SERCA2b reveal the mechanism of regulation by the luminal extension tail.
Sci Adv, 6, 2020
|
|
