5EQ7
 
 | |
5EQA
 
 | |
2QT4
 
 | | Atomic-resolution crystal structure of the natural form of Scytovirin | | Descriptor: | scytovirin | | Authors: | Moulaei, T, Botos, I, Ziolkowska, N.E, Dauter, Z, Wlodawer, A. | | Deposit date: | 2007-08-01 | | Release date: | 2007-11-27 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Atomic-resolution crystal structure of the antiviral lectin scytovirin.
Protein Sci., 16, 2007
|
|
5EQ9
 
 | |
5EME
 
 | | Complex of RNA r(GCAGCAGC) with antisense PNA p(CTGCTGC) | | Descriptor: | Antisense PNA strand, CHLORIDE ION, RNA (5'-R(*GP*CP*AP*GP*CP*AP*GP*C)-3') | | Authors: | Kiliszek, A, Banaszak, K, Dauter, Z, Rypniewski, W. | | Deposit date: | 2015-11-06 | | Release date: | 2016-01-13 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | The first crystal structures of RNA-PNA duplexes and a PNA-PNA duplex containing mismatches-toward anti-sense therapy against TREDs.
Nucleic Acids Res., 44, 2016
|
|
2PVA
 
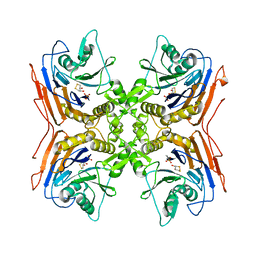 | | OXIDIZED PENICILLIN V ACYLASE FROM B. SPHAERICUS | | Descriptor: | DITHIANE DIOL, PENICILLIN V ACYLASE | | Authors: | Suresh, C.G, Pundle, A.V, Rao, K.N, SivaRaman, H, Brannigan, J.A, McVey, C.E, Verma, C.S, Dauter, Z, Dodson, E.J, Dodson, G.G. | | Deposit date: | 1998-11-13 | | Release date: | 2000-07-26 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Penicillin V acylase crystal structure reveals new Ntn-hydrolase family members.
Nat.Struct.Biol., 6, 1999
|
|
2QSK
 
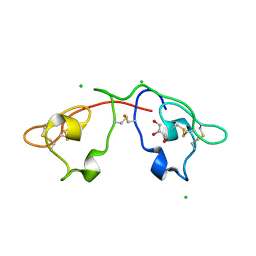 | | Atomic-resolution crystal structure of the Recombinant form of Scytovirin | | Descriptor: | CHLORIDE ION, GLYCEROL, scytovirin | | Authors: | Moulaei, T, Botos, I, Ziolkowska, N.E, Dauter, Z, Wlodawer, A. | | Deposit date: | 2007-07-31 | | Release date: | 2007-11-27 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Atomic-resolution crystal structure of the antiviral lectin scytovirin.
Protein Sci., 16, 2007
|
|
6M9D
 
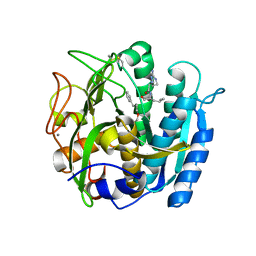 | | PSEUDOMONAS SERINE-CARBOXYL PROTEINASE (SEDOLISIN) COMPLEXED WITH THE INHIBITOR Chymostatin | | Descriptor: | CALCIUM ION, Chymostatin A, SEDOLISIN | | Authors: | Wlodawer, A, Li, M, Gustchina, A, Dauter, Z, Uchida, K, Oyama, H, Goldfarb, N.E, Dunn, B.M, Oda, K. | | Deposit date: | 2018-08-23 | | Release date: | 2018-10-24 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Inhibitor complexes of the Pseudomonas serine-carboxyl proteinase
Biochemistry, 40, 2001
|
|
6M9F
 
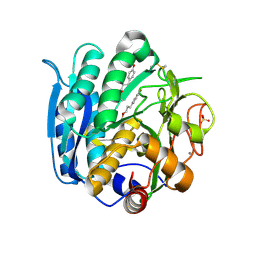 | | PSEUDOMONAS SERINE-CARBOXYL PROTEINASE (SEDOLISIN) COMPLEXED WITH THE INHIBITOR Tyrostatin | | Descriptor: | CALCIUM ION, SEDOLISIN, SULFATE ION, ... | | Authors: | Wlodawer, A, Li, M, Gustchina, A, Dauter, Z, Uchida, K, Oyama, H, Goldfarb, N.E, Dunn, B.M, Oda, K. | | Deposit date: | 2018-08-23 | | Release date: | 2018-10-24 | | Last modified: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Inhibitor complexes of the Pseudomonas serine-carboxyl proteinase
Biochemistry, 40, 2001
|
|
6M8Y
 
 | | PSEUDOMONAS SERINE-CARBOXYL PROTEINASE (SEDOLISIN) COMPLEXED WITH THE INHIBITOR AIPF | | Descriptor: | AIPF PEPTIDE INHIBITOR, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Wlodawer, A, Li, M, Gustchina, A, Dauter, Z, Uchida, K, Oyama, H, Goldfarb, N.E, Dunn, B.M, Oda, K. | | Deposit date: | 2018-08-22 | | Release date: | 2018-10-24 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Inhibitor complexes of the Pseudomonas serine-carboxyl proteinase
Biochemistry, 40, 2001
|
|
6M8W
 
 | | PSEUDOMONAS SERINE-CARBOXYL PROTEINASE (SEDOLISIN) COMPLEXED WITH THE INHIBITOR AIAF | | Descriptor: | AIAF PEPTIDE INHIBITOR, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Wlodawer, A, Li, M, Gustchina, A, Dauter, Z, Uchida, K, Oyama, H, Goldfarb, N.E, Dunn, B.M, Oda, K. | | Deposit date: | 2018-08-22 | | Release date: | 2018-10-24 | | Last modified: | 2025-04-02 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Inhibitor complexes of the Pseudomonas serine-carboxyl proteinase
Biochemistry, 40, 2001
|
|
5VEH
 
 | | Re-refinement OF THE PDB STRUCTURE 1yiz of Aedes aegypti kynurenine aminotransferase | | Descriptor: | BROMIDE ION, GLYCEROL, Kynurenine aminotransferase | | Authors: | Wlodawer, A, Dauter, Z, Minor, W, Stanfield, R, Porebski, P, Jaskolski, M, Pozharski, E, Weichenberger, C.X, Rupp, B. | | Deposit date: | 2017-04-04 | | Release date: | 2017-11-29 | | Last modified: | 2022-04-13 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Detect, correct, retract: How to manage incorrect structural models.
FEBS J., 285, 2018
|
|
5VEQ
 
 | | MOUSE KYNURENINE AMINOTRANSFERASE III, RE-REFINEMENT OF THE PDB STRUCTURE 3E2Y | | Descriptor: | 4'-DEOXY-4'-AMINOPYRIDOXAL-5'-PHOSPHATE, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CALCIUM ION, ... | | Authors: | Wlodawer, A, Dauter, Z, Minor, W, Stanfield, R, Porebski, P, Jaskolski, M, Pozharski, E, Weichenberger, C.X, Rupp, B. | | Deposit date: | 2017-04-05 | | Release date: | 2017-11-29 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Detect, correct, retract: How to manage incorrect structural models.
FEBS J., 285, 2018
|
|
5VF2
 
 | | scFv 2D10 re-refined as a complex with trehalose replacing the original alpha-1,6-mannobiose | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, MAGNESIUM ION, UNKNOWN ATOM OR ION, ... | | Authors: | Porebski, P.J, Wlodawer, A, Dauter, Z, Minor, W, Stanfield, R, Jaskolski, M, Pozharski, E, Weichenberger, C.X, Rupp, B. | | Deposit date: | 2017-04-06 | | Release date: | 2017-12-06 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Detect, correct, retract: How to manage incorrect structural models.
FEBS J., 285, 2018
|
|
5VLC
 
 | |
5VGA
 
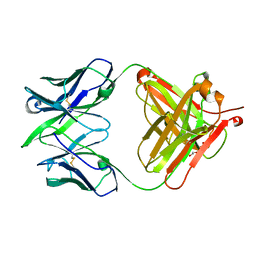 | | Alternative model for Fab 36-65 | | Descriptor: | Fab 36-65 heavy chain, Fab 36-65 light chain, TRIETHYLENE GLYCOL | | Authors: | Stanfield, R.L, Rupp, B, Wlodawer, A, Dauter, Z, Porebski, P.J, Minor, W, Jaskolski, M, Pozharski, E, Weichenberger, C.X. | | Deposit date: | 2017-04-10 | | Release date: | 2017-12-06 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Detect, correct, retract: How to manage incorrect structural models.
FEBS J., 285, 2018
|
|
5VET
 
 | | PHOSPHOLIPASE A2, RE-REFINEMENT OF THE PDB STRUCTURE 1JQ8 WITHOUT THE PUTATIVE COMPLEXED OLIGOPEPTIDE | | Descriptor: | Phospholipase A2 VRV-PL-VIIIa | | Authors: | Wlodawer, A, Dauter, Z, Minor, W, Stanfield, R, Porebski, P, Jaskolski, M, Pozharski, E, Weichenberger, C.X, Rupp, B. | | Deposit date: | 2017-04-05 | | Release date: | 2018-06-20 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Detect, correct, retract: How to manage incorrect structural models.
FEBS J., 285, 2018
|
|
6NIC
 
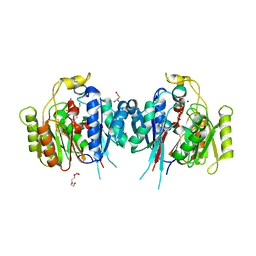 | | Crystal Structure of Medicago truncatula Agmatine Iminohydrolase (Deiminase) in Complex with 6-aminohexanamide | | Descriptor: | 1,2-ETHANEDIOL, 6-aminohexanamide, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Sekula, B, Dauter, Z. | | Deposit date: | 2018-12-27 | | Release date: | 2019-03-20 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural Study of Agmatine Iminohydrolase FromMedicago truncatula, the Second Enzyme of the Agmatine Route of Putrescine Biosynthesis in Plants.
Front Plant Sci, 10, 2019
|
|
3PVA
 
 | | PENICILLIN V ACYLASE FROM B. SPHAERICUS | | Descriptor: | PROTEIN (PENICILLIN V ACYLASE) | | Authors: | Suresh, C.G, Pundle, A.V, Rao, K.N, Sivaraman, H, Brannigan, J.A, Mcvey, C.E, Verma, C.S, Dauter, Z, Dodson, E.J, Dodson, G.G. | | Deposit date: | 1998-11-13 | | Release date: | 1999-11-15 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Penicillin V acylase crystal structure reveals new Ntn-hydrolase family members.
Nat.Struct.Biol., 6, 1999
|
|
5VER
 
 | | MOUSE KYNURENINE AMINOTRANSFERASE III, RE-REFINEMENT OF THE PDB STRUCTURE 3E2Z | | Descriptor: | 4'-DEOXY-4'-AMINOPYRIDOXAL-5'-PHOSPHATE, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CALCIUM ION, ... | | Authors: | Wlodawer, A, Dauter, Z, Minor, W, Stanfield, R, Porebski, P, Jaskolski, M, Pozharski, E, Weichenberger, C.X, Rupp, B. | | Deposit date: | 2017-04-05 | | Release date: | 2017-11-29 | | Last modified: | 2022-04-13 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | Detect, correct, retract: How to manage incorrect structural models.
FEBS J., 285, 2018
|
|
5VLD
 
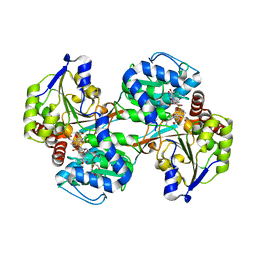 | |
6O65
 
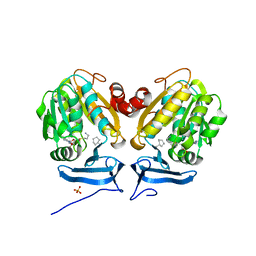 | |
6NIB
 
 | | Crystal Structure of Medicago truncatula Agmatine Iminohydrolase (Deiminase) | | Descriptor: | DI(HYDROXYETHYL)ETHER, GLYCEROL, Porphyromonas-type peptidyl-arginine deiminase, ... | | Authors: | Sekula, B, Dauter, Z. | | Deposit date: | 2018-12-27 | | Release date: | 2019-03-20 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Structural Study of Agmatine Iminohydrolase FromMedicago truncatula, the Second Enzyme of the Agmatine Route of Putrescine Biosynthesis in Plants.
Front Plant Sci, 10, 2019
|
|
6B91
 
 | | Crystal structure of the N-terminal domain of human METTL16 | | Descriptor: | 1,2-ETHANEDIOL, SODIUM ION, U6 small nuclear RNA (adenine-(43)-N(6))-methyltransferase | | Authors: | Ruszkowska, A, Ruszkowski, M, Dauter, Z, Brown, J.A. | | Deposit date: | 2017-10-09 | | Release date: | 2018-04-04 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Structural insights into the RNA methyltransferase domain of METTL16.
Sci Rep, 8, 2018
|
|
5VEP
 
 | | MOUSE KYNURENINE AMINOTRANSFERASE III, RE-REFINEMENT OF THE PDB STRUCTURE 3E2F | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CALCIUM ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Wlodawer, A, Dauter, Z, Minor, W, Stanfield, R, Porebski, P, Jaskolski, M, Pozharski, E, Weichenberger, C.X, Rupp, B. | | Deposit date: | 2017-04-05 | | Release date: | 2017-11-29 | | Last modified: | 2022-04-13 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Detect, correct, retract: How to manage incorrect structural models.
FEBS J., 285, 2018
|
|
