3FO3
 
 | | Structure of the Thioalkalivibrio nitratireducens cytochrome c nitrite reductase reduced by sodium dithionite (sulfite complex) | | Descriptor: | 1-(2-METHOXY-ETHOXY)-2-{2-[2-(2-METHOXY-ETHOXY]-ETHOXY}-ETHANE, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ACETATE ION, ... | | Authors: | Trofimov, A.A, Polyakov, K.M, Boyko, K.M, Slutsky, A, Tikhonova, T.V, Antipov, A.N, Zvyagilskaya, R.A, Popov, A.N, Lamzin, V.S, Bourenkov, G.P, Popov, V.O. | | Deposit date: | 2008-12-27 | | Release date: | 2009-12-29 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structures of complexes of octahaem cytochrome c nitrite reductase from Thioalkalivibrio nitratireducens with sulfite and cyanide
Acta Crystallogr.,Sect.D, 66, 2010
|
|
8ONO
 
 | | Modified oligopeptidase B from S. proteamaculans in intermediate conformation with 5 spermine molecule at 1.65 A resolution | | Descriptor: | Oligopeptidase B, SPERMINE | | Authors: | Petrenko, D.E, Boyko, K.M, Nikolaeva, A.Y, Vlaskina, A.V, Mikhailova, A.G, Timofeev, V.I, Rakitina, T.V. | | Deposit date: | 2023-04-03 | | Release date: | 2023-05-31 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Modified oligopeptidase B from S. proteomaculans in intermediate conformation with 5 spermine molecule at 1.65 A resolution
To Be Published
|
|
5JFQ
 
 | | Geranylgeranyl Pyrophosphate Synthetase from archaeon Geoglobus acetivorans | | Descriptor: | Geranylgeranyl Pyrophosphate Synthetase | | Authors: | Petrova, T, Boyko, K.M, Nikolaeva, A.Y, Stekhanova, T.N, Mardanov, A.V, Rakitin, A.L, Ravin, N.V, Popov, V.O. | | Deposit date: | 2016-04-19 | | Release date: | 2017-05-24 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Structural characterization of geranylgeranyl pyrophosphate synthase GACE1337 from the hyperthermophilic archaeon Geoglobus acetivorans.
Extremophiles, 22, 2018
|
|
8QPT
 
 | | Crystal structure of pyrophosphatase from Ogataea parapolymorpha | | Descriptor: | GLYCEROL, MAGNESIUM ION, inorganic diphosphatase | | Authors: | Matyuta, I.O, Rodina, E.V, Vorobyeva, N.N, Kurilova, S.A, Bezpalaya, E.Y, Boyko, K.M. | | Deposit date: | 2023-10-03 | | Release date: | 2024-09-04 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The crystal structure of yeast mitochondrial type pyrophosphatase provides a model to study pathological mutations in its human ortholog.
Biochem.Biophys.Res.Commun., 738, 2024
|
|
8RAF
 
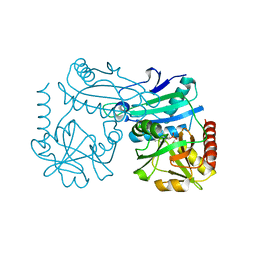 | | Crystal structure of D-amino acid transaminase from Haliscomenobacter hydrossis point mutant R90I (holo form) | | Descriptor: | Aminotransferase class IV, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Matyuta, I.O, Bakunova, A.K, Minyaev, M.E, Popov, V.O, Bezsudnova, E.Y, Boyko, K.M. | | Deposit date: | 2023-12-01 | | Release date: | 2023-12-27 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Multifunctionality of arginine residues in the active sites of non-canonical d-amino acid transaminases.
Arch.Biochem.Biophys., 756, 2024
|
|
8RAI
 
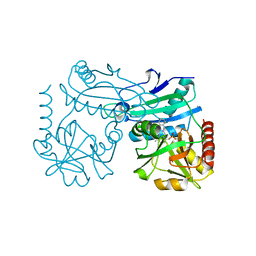 | | Crystal structure of D-amino acid transaminase from Haliscomenobacter hydrossis point mutant R90I complexed with phenylhydrazine | | Descriptor: | Aminotransferase class IV, GLYCEROL, [6-methyl-5-oxidanyl-4-[(2-phenylhydrazinyl)methyl]pyridin-3-yl]methyl dihydrogen phosphate | | Authors: | Matyuta, I.O, Bakunova, A.K, Minyaev, M.E, Popov, V.O, Bezsudnova, E.Y, Boyko, K.M. | | Deposit date: | 2023-12-01 | | Release date: | 2023-12-27 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Multifunctionality of arginine residues in the active sites of non-canonical d-amino acid transaminases.
Arch.Biochem.Biophys., 756, 2024
|
|
6FP5
 
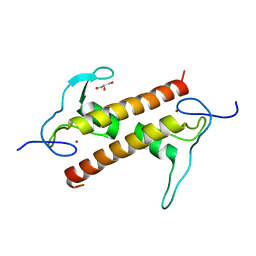 | | Crystal structure of ZAD-domain of CG2712 protein from D.melanogaster | | Descriptor: | CG2712, GLYCEROL, ZINC ION | | Authors: | Boyko, K.M, Nikolaeva, A.Y, Bonchuk, A.N, Kachalova, G.S, Georgiev, P.G, Popov, V.O. | | Deposit date: | 2018-02-09 | | Release date: | 2019-08-21 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis of diversity and homodimerization specificity of zinc-finger-associated domains in Drosophila.
Nucleic Acids Res., 49, 2021
|
|
7O45
 
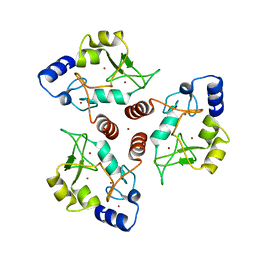 | | Crystal structure of ADD domain of the human DNMT3B methyltransferase | | Descriptor: | BROMIDE ION, Isoform 6 of DNA (cytosine-5)-methyltransferase 3B, ZINC ION | | Authors: | Boyko, K.M, Nikolaeva, A.Y, Bonchuk, A.N, Georgiev, P.G, Popov, V.O. | | Deposit date: | 2021-04-05 | | Release date: | 2022-04-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of the DNMT3B ADD domain suggests the absence of a DNMT3A-like autoinhibitory mechanism.
Biochem.Biophys.Res.Commun., 619, 2022
|
|
3GM6
 
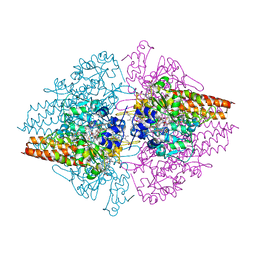 | | Structure of the Thioalkalivibrio nitratireducens cytochrome c nitrite reductase in complex with phosphate | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, CALCIUM ION, ... | | Authors: | Trofimov, A.A, Polyakov, K.M, Boyko, K.M, Filimonenkov, A.A, Dorovatovsky, P.V, Tikhonova, T.V, Popov, V.O. | | Deposit date: | 2009-03-13 | | Release date: | 2009-08-04 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of octaheme cytochrome c nitrite reductase from Thioalkalivibrio nitratireducens in a complex with phosphate
Crystallography Reports, 55, 2010
|
|
3SXQ
 
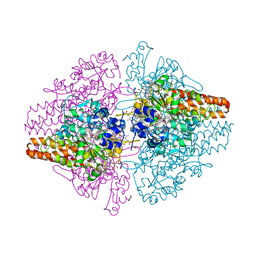 | | Structure of a hexameric multiheme c nitrite reductase from the extremophile bacterium Thiolkalivibrio paradoxus | | Descriptor: | CALCIUM ION, CHLORIDE ION, COBALT (II) ION, ... | | Authors: | Polyakov, K.M, Trofimov, A.A, Tikhonova, T.V, Tikhonov, A.V, Boyko, K.M, Popov, V.O. | | Deposit date: | 2011-07-15 | | Release date: | 2012-09-26 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Comparative structural and functional analysis of two octaheme nitrite reductases from closely related Thioalkalivibrio species.
Febs J., 279, 2012
|
|
4RGZ
 
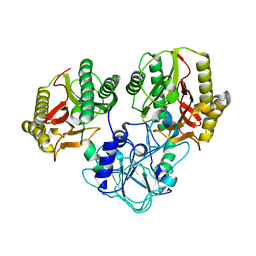 | | Crystal structure of recombinant prolidase from Thermococcus sibiricus at P21221 spacegroup | | Descriptor: | PHOSPHATE ION, Xaa-Pro aminopeptidase, ZINC ION | | Authors: | Timofeev, V.I, Korgenevsky, D.A, Gorbacheva, M.A, Boyko, K.M, Slutsky, E, Rakitina, T.V, Lipkin, A.V, Popov, V.O. | | Deposit date: | 2014-10-01 | | Release date: | 2015-08-12 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of recombinant prolidase from Thermococcus sibiricus in space group P21221.
Acta Crystallogr.,Sect.F, 71, 2015
|
|
3UU9
 
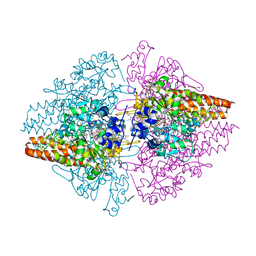 | | Structure of the free TvNiRb form of Thioalkalivibrio nitratireducens cytochrome c nitrite reductase | | Descriptor: | CALCIUM ION, Eight-heme nitrite reductase, HEME C, ... | | Authors: | Trofimov, A.A, Polyakov, K.M, Tikhonova, T.V, Tikhonov, A.V, Dorovatovskii, P.V, Popov, V.O. | | Deposit date: | 2011-11-28 | | Release date: | 2012-01-25 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Covalent modifications of the catalytic tyrosine in octahaem cytochrome c nitrite reductase and their effect on the enzyme activity.
Acta Crystallogr.,Sect.D, 68, 2012
|
|
3RKH
 
 | | Structure of the Thioalkalivibrio nitratireducens cytochrome c nitrite reductase in a complex with nitrite (full occupancy) | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, CALCIUM ION, Eight-heme nitrite reductase, ... | | Authors: | Trofimov, A.A, Polyakov, K.M, Tikhonova, T.V, Tikhonov, A.V, Dorovatovskii, P.V, Popov, V.O. | | Deposit date: | 2011-04-18 | | Release date: | 2011-06-15 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Covalent modifications of the catalytic tyrosine in octahaem cytochrome c nitrite reductase and their effect on the enzyme activity.
Acta Crystallogr.,Sect.D, 68, 2012
|
|
3OWM
 
 | | Structure of the Thioalkalivibrio nitratireducens cytochrome c nitrite reductase in a complex with hydroxylamine | | Descriptor: | 1-(2-METHOXY-ETHOXY)-2-{2-[2-(2-METHOXY-ETHOXY]-ETHOXY}-ETHANE, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CALCIUM ION, ... | | Authors: | Trofimov, A.A, Polyakov, K.M, Boyko, K.M, Tikhonova, T.V, Lamzin, V.S, Bourenkov, G.P, Popov, V.O. | | Deposit date: | 2010-09-20 | | Release date: | 2011-10-26 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Binding of sulfite by the Thioalkalivibrio nitratireducens cytochrome c nitrite reductase
To be Published
|
|
5MR0
 
 | | Thermophilic archaeal branched-chain amino acid transaminases from Geoglobus acetivorans and Archaeoglobus fulgidus: biochemical and structural characterisation | | Descriptor: | 1,2-ETHANEDIOL, 3-[O-PHOSPHONOPYRIDOXYL]--AMINO-BENZOIC ACID, CHLORIDE ION, ... | | Authors: | Isupov, M.N, Littlechild, J.A, James, P, Sayer, C, Sutter, J.M, Schmidt, M, Schoenheit, P. | | Deposit date: | 2016-12-21 | | Release date: | 2018-01-17 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Thermostable Branched-Chain Amino Acid Transaminases From the ArchaeaGeoglobus acetivoransandArchaeoglobus fulgidus: Biochemical and Structural Characterization.
Front Bioeng Biotechnol, 7, 2019
|
|
5MQZ
 
 | | Archaeal branched-chain amino acid aminotransferase from Archaeoglobus fulgidus; holoform | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | James, P, Isupov, M.N, Sayer, C, Littlechild, J.A, Sutter, J.M, Schmidt, M, Schoenheit, P. | | Deposit date: | 2016-12-21 | | Release date: | 2018-01-17 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Thermostable Branched-Chain Amino Acid Transaminases From the Archaea Geoglobus acetivorans and Archaeoglobus fulgidus : Biochemical and Structural Characterization.
Front Bioeng Biotechnol, 7, 2019
|
|
6UWE
 
 | | Crystal structure of recombinant thiocyanate dehydrogenase from Thioalkalivibrio paradoxus saturated with copper | | Descriptor: | COPPER (II) ION, UNKNOWN ATOM OR ION, thiocyanate dehydrogenase | | Authors: | Shabalin, I.G, Osipov, E, Tikhonova, T.V, Rakitina, T.V, Boyko, K.M, Popov, V.O. | | Deposit date: | 2019-11-05 | | Release date: | 2019-11-27 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Trinuclear copper biocatalytic center forms an active site of thiocyanate dehydrogenase.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
3F29
 
 | | Structure of the Thioalkalivibrio nitratireducens cytochrome c nitrite reductase in complex with sulfite | | Descriptor: | 1-(2-METHOXY-ETHOXY)-2-{2-[2-(2-METHOXY-ETHOXY]-ETHOXY}-ETHANE, CALCIUM ION, Eight-heme nitrite reductase, ... | | Authors: | Trofimov, A.A, Polyakov, K.M, Boyko, K.M, Slutsky, A, Tikhonova, T.V, Antipov, A.N, Zvyagilskaya, R.A, Popov, A.N, Lamzin, V.S, Bourenkov, G.P, Popov, V.O. | | Deposit date: | 2008-10-29 | | Release date: | 2008-12-09 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Binding of sulfite by the Thioalkalivibrio nitratireducens cytochrome c nitrite reductase
To be Published
|
|
8RC6
 
 | |
