3DMK
 
 | | Crystal structure of Down Syndrome Cell Adhesion Molecule (DSCAM) isoform 1.30.30, N-terminal eight Ig domains | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Down Syndrome Cell Adhesion Molecule (DSCAM) isoform 1.30.30, ... | | Authors: | Sawaya, M.R, Wojtowicz, W.M, Eisenberg, D, Zipursky, S.L. | | Deposit date: | 2008-07-01 | | Release date: | 2008-10-07 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (4.19 Å) | | Cite: | A double S shape provides the structural basis for the extraordinary binding specificity of Dscam isoforms.
Cell(Cambridge,Mass.), 134, 2008
|
|
6LOH
 
 | |
7WTK
 
 | | SARS-CoV-2 Omicron variant spike in complex with Fab XGv286 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of XGv286, ... | | Authors: | Wang, X, Fu, W. | | Deposit date: | 2022-02-04 | | Release date: | 2022-12-14 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Selection and structural bases of potent broadly neutralizing antibodies from 3-dose vaccinees that are highly effective against diverse SARS-CoV-2 variants, including Omicron sublineages.
Cell Res., 32, 2022
|
|
7WTJ
 
 | | SARS-CoV-2 Omicron variant spike RBD in complex with Fab XGv286 | | Descriptor: | Heavy chain of XGv286, Light chain of XGv286, Spike protein S1 | | Authors: | Wang, X, Fu, W. | | Deposit date: | 2022-02-04 | | Release date: | 2022-12-14 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Selection and structural bases of potent broadly neutralizing antibodies from 3-dose vaccinees that are highly effective against diverse SARS-CoV-2 variants, including Omicron sublineages.
Cell Res., 32, 2022
|
|
7WTG
 
 | | SARS-CoV-2 Omicron variant spike RBD in complex with Fab XGv051 | | Descriptor: | Heavy chain of XGv051, Light chain of XGv051, Spike protein S1 | | Authors: | Wang, X, Fu, W. | | Deposit date: | 2022-02-04 | | Release date: | 2022-12-14 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Selection and structural bases of potent broadly neutralizing antibodies from 3-dose vaccinees that are highly effective against diverse SARS-CoV-2 variants, including Omicron sublineages.
Cell Res., 32, 2022
|
|
7WTH
 
 | | SARS-CoV-2 Omicron variant spike RBD in complex with Fab XGv264 | | Descriptor: | Heavy chain of XGv264, Light chain of XGv264, Spike protein S1 | | Authors: | Wang, X, Fu, W. | | Deposit date: | 2022-02-04 | | Release date: | 2022-12-14 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Selection and structural bases of potent broadly neutralizing antibodies from 3-dose vaccinees that are highly effective against diverse SARS-CoV-2 variants, including Omicron sublineages.
Cell Res., 32, 2022
|
|
7WTF
 
 | | SARS-CoV-2 Omicron variant spike in complex with Fab XGv051 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of XGv051, ... | | Authors: | Wang, X, Fu, W. | | Deposit date: | 2022-02-04 | | Release date: | 2022-12-14 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Selection and structural bases of potent broadly neutralizing antibodies from 3-dose vaccinees that are highly effective against diverse SARS-CoV-2 variants, including Omicron sublineages.
Cell Res., 32, 2022
|
|
7WTI
 
 | | SARS-CoV-2 Omicron variant spike in complex with Fab XGv264 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of XGv264, Light chain of XGv264, ... | | Authors: | Wang, X, Fu, W. | | Deposit date: | 2022-02-04 | | Release date: | 2022-12-21 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Selection and structural bases of potent broadly neutralizing antibodies from 3-dose vaccinees that are highly effective against diverse SARS-CoV-2 variants, including Omicron sublineages.
Cell Res., 32, 2022
|
|
3MZ8
 
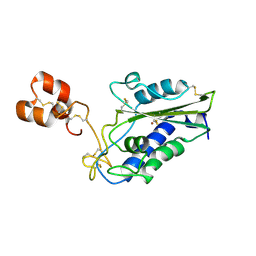 | | Crystal Structure of Zinc-Bound Natrin From Naja atra | | Descriptor: | Natrin-1, ZINC ION | | Authors: | Wang, Y.L, Hsieh, Y.C, Liu, J.S, Chen, C.J, Wu, W.G. | | Deposit date: | 2010-05-12 | | Release date: | 2010-09-29 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Cobra CRISP functions as an inflammatory modulator via a novel Zn2+- and heparan sulfate- dependent transcriptional regulation of endothelial cell adhesion molecules
J.Biol.Chem., 285, 2010
|
|
3OJI
 
 | | X-ray crystal structure of the Py13 -pyrabactin complex | | Descriptor: | 4-bromo-N-(pyridin-2-ylmethyl)naphthalene-1-sulfonamide, Abscisic acid receptor PYL3, SULFATE ION | | Authors: | Zhang, X, Zhang, Q, Wang, G, Chen, Z. | | Deposit date: | 2010-08-23 | | Release date: | 2011-08-10 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Complex Structures of the Abscisic Acid Receptor PYL3/RCAR13 Reveal a Unique Regulatory Mechanism
Structure, 20, 2012
|
|
8GTH
 
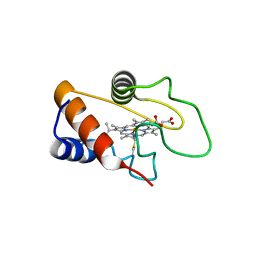 | |
7RGW
 
 | |
3TYV
 
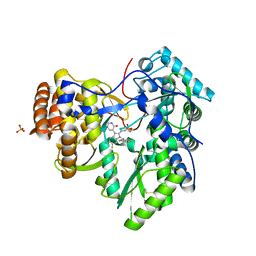 | |
7XPO
 
 | | Crystal Structure of UDP-Glc/GlcNAc 4-Epimerase with NAD/UDP-Glc | | Descriptor: | GLYCEROL, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, UDP-glucose 4-epimerase, ... | | Authors: | Chen, Y.H, Wang, X.C, Zhang, C.R. | | Deposit date: | 2022-05-05 | | Release date: | 2023-05-17 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | A maize epimerase modulates cell wall synthesis and glycosylation during stomatal morphogenesis.
Nat Commun, 14, 2023
|
|
7XPQ
 
 | |
7XPP
 
 | |
5YYL
 
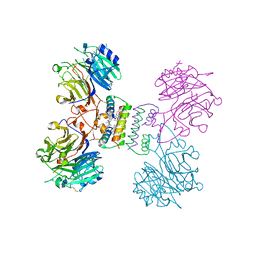 | | Structure of Major Royal Jelly Protein 1 Oligomer | | Descriptor: | (3beta,14beta,17alpha)-ergosta-5,24(28)-dien-3-ol, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Tian, W, Chen, Z. | | Deposit date: | 2017-12-10 | | Release date: | 2018-08-08 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Architecture of the native major royal jelly protein 1 oligomer.
Nat Commun, 9, 2018
|
|
6OSY
 
 | |
6OT1
 
 | |
7V97
 
 | | Arsenic-bound p53 DNA-binding domain mutant V272M | | Descriptor: | ARSENIC, Cellular tumor antigen p53, ZINC ION | | Authors: | Lu, M, Xing, Y.F, Wang, Z.Y, Ni, Y, Song, H.X. | | Deposit date: | 2021-08-24 | | Release date: | 2022-08-31 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Diverse rescue potencies of p53 mutations to ATO are predetermined by intrinsic mutational properties.
Sci Transl Med, 15, 2023
|
|
4DSB
 
 | | Complex Structure of Abscisic Acid Receptor PYL3 with (+)-ABA in Spacegroup of I 212121 at 2.70A | | Descriptor: | (2Z,4E)-5-[(1S)-1-hydroxy-2,6,6-trimethyl-4-oxocyclohex-2-en-1-yl]-3-methylpenta-2,4-dienoic acid, Abscisic acid receptor PYL3 | | Authors: | Zhang, X, Zhang, Q, Chen, Z. | | Deposit date: | 2012-02-18 | | Release date: | 2012-06-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Complex Structures of the Abscisic Acid Receptor PYL3/RCAR13 Reveal a Unique Regulatory Mechanism
Structure, 20, 2012
|
|
4DSC
 
 | | Complex structure of abscisic acid receptor PYL3 with (+)-ABA in spacegroup of H32 at 1.95A | | Descriptor: | (2Z,4E)-5-[(1S)-1-hydroxy-2,6,6-trimethyl-4-oxocyclohex-2-en-1-yl]-3-methylpenta-2,4-dienoic acid, Abscisic acid receptor PYL3, MAGNESIUM ION | | Authors: | Zhang, X, Chen, Z. | | Deposit date: | 2012-02-18 | | Release date: | 2012-06-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Complex Structures of the Abscisic Acid Receptor PYL3/RCAR13 Reveal a Unique Regulatory Mechanism
Structure, 20, 2012
|
|
8HI5
 
 | |
8HI6
 
 | |
4DS8
 
 | | Complex structure of abscisic acid receptor PYL3-(+)-ABA-HAB1 in the presence of Mn2+ | | Descriptor: | (2Z,4E)-5-[(1S)-1-hydroxy-2,6,6-trimethyl-4-oxocyclohex-2-en-1-yl]-3-methylpenta-2,4-dienoic acid, Abscisic acid receptor PYL3, GLYCEROL, ... | | Authors: | Zhang, X, Zhang, Q, Wang, G, Chen, Z. | | Deposit date: | 2012-02-18 | | Release date: | 2012-06-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Complex Structures of the Abscisic Acid Receptor PYL3/RCAR13 Reveal a Unique Regulatory Mechanism
Structure, 20, 2012
|
|
