3WG4
 
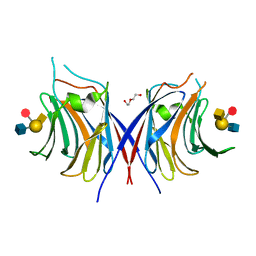 | | Crystal structure of Agrocybe cylindracea galectin mutant (N46A) with blood type A antigen tetraose | | Descriptor: | DI(HYDROXYETHYL)ETHER, Galactoside-binding lectin, alpha-L-fucopyranose-(1-2)-[2-acetamido-2-deoxy-alpha-D-galactopyranose-(1-3)]beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Kuwabara, N, Hu, D, Tateno, H, Makio, H, Hirabayashi, J, Kato, R. | | Deposit date: | 2013-07-25 | | Release date: | 2013-11-27 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Conformational change of a unique sequence in a fungal galectin from Agrocybe cylindracea controls glycan ligand-binding specificity.
Febs Lett., 587, 2013
|
|
3WAM
 
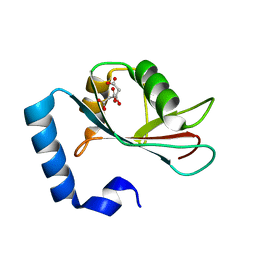 | | Crystal structure of human LC3C_8-125 | | Descriptor: | CITRIC ACID, Microtubule-associated proteins 1A/1B light chain 3C | | Authors: | Suzuki, H, Tabata, K, Morita, E, Kawasaki, M, Kato, R, Dobson, R.C.J, Yoshimori, T, Wakatsuki, S. | | Deposit date: | 2013-05-06 | | Release date: | 2013-12-25 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural basis of the autophagy-related LC3/Atg13 LIR complex: recognition and interaction mechanism.
Structure, 22, 2014
|
|
3WG3
 
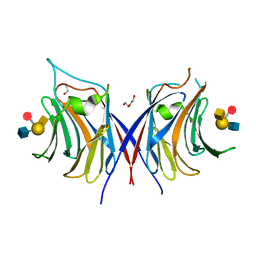 | | Crystal structure of Agrocybe cylindracea galectin with blood type A antigen tetraose | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, Galactoside-binding lectin, ... | | Authors: | Kuwabara, N, Hu, D, Tateno, H, Makio, H, Hirabayashi, J, Kato, R. | | Deposit date: | 2013-07-25 | | Release date: | 2013-11-27 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Conformational change of a unique sequence in a fungal galectin from Agrocybe cylindracea controls glycan ligand-binding specificity.
Febs Lett., 587, 2013
|
|
3WD7
 
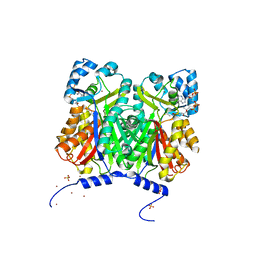 | | Type III polyketide synthase | | Descriptor: | COENZYME A, NICKEL (II) ION, SULFATE ION, ... | | Authors: | Mori, T, Shimokawa, Y, Matsui, T, Kato, R, Sugio, S, Morita, H, Abe, I. | | Deposit date: | 2013-06-10 | | Release date: | 2013-09-04 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Cloning, characterization, and crystal structure analysis of novel type III polyketide synthases from Citrus microcarpa
To be Published
|
|
3WAP
 
 | | Crystal structure of Atg13 LIR-fused human LC3C_8-125 | | Descriptor: | Autophagy-related protein 13, Microtubule-associated proteins 1A/1B light chain 3C | | Authors: | Suzuki, H, Tabata, K, Morita, E, Kawasaki, M, Kato, R, Dobson, R.C.J, Yoshimori, T, Wakatsuki, S. | | Deposit date: | 2013-05-06 | | Release date: | 2013-12-25 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural basis of the autophagy-related LC3/Atg13 LIR complex: recognition and interaction mechanism.
Structure, 22, 2014
|
|
3WG2
 
 | | Crystal structure of Agrocybe cylindracea galectin mutant (N46A) | | Descriptor: | Galactoside-binding lectin, TRIETHYLENE GLYCOL | | Authors: | Kuwabara, N, Hu, D, Tateno, H, Makio, H, Hirabayashi, J, Kato, R. | | Deposit date: | 2013-07-25 | | Release date: | 2013-11-27 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Conformational change of a unique sequence in a fungal galectin from Agrocybe cylindracea controls glycan ligand-binding specificity.
Febs Lett., 587, 2013
|
|
3WAL
 
 | | Crystal structure of human LC3A_2-121 | | Descriptor: | D-MALATE, Microtubule-associated proteins 1A/1B light chain 3A | | Authors: | Suzuki, H, Tabata, K, Morita, E, Kawasaki, M, Kato, R, Dobson, R.C.J, Yoshimori, T, Wakatsuki, S. | | Deposit date: | 2013-05-06 | | Release date: | 2013-12-25 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis of the autophagy-related LC3/Atg13 LIR complex: recognition and interaction mechanism.
Structure, 22, 2014
|
|
3WG1
 
 | | Crystal structure of Agrocybe cylindracea galectin with lactose | | Descriptor: | Galactoside-binding lectin, beta-D-galactopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Kuwabara, N, Hu, D, Tateno, H, Makio, H, Hirabayashi, J, Kato, R. | | Deposit date: | 2013-07-25 | | Release date: | 2013-11-27 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Conformational change of a unique sequence in a fungal galectin from Agrocybe cylindracea controls glycan ligand-binding specificity.
Febs Lett., 587, 2013
|
|
3WLU
 
 | | Crystal Structure of human galectin-9 NCRD with Selenolactose | | Descriptor: | 2-(trimethylsilyl)ethyl 4-O-beta-D-galactopyranosyl-6-Se-methyl-6-seleno-beta-D-glucopyranoside, Galectin-9 | | Authors: | Makyio, H, Suzuki, T, Ando, H, Yamada, Y, Ishida, H, Kiso, M, Wakatsuki, S, Kato, R. | | Deposit date: | 2013-11-14 | | Release date: | 2014-11-05 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Expanded potential of seleno-carbohydrates as a molecular tool for X-ray structural determination of a carbohydrate-protein complex with single/multi-wavelength anomalous dispersion phasing
Bioorg.Med.Chem., 22, 2014
|
|
6L7U
 
 | | Crystal structure of FKRP in complex with Ba ion, Ba-SAD data | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, BARIUM ION, Fukutin-related protein, ... | | Authors: | Kuwabara, N. | | Deposit date: | 2019-11-03 | | Release date: | 2020-01-15 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Crystal structures of fukutin-related protein (FKRP), a ribitol-phosphate transferase related to muscular dystrophy.
Nat Commun, 11, 2020
|
|
6L7T
 
 | | Crystal structure of FKRP in complex with Mg ion, Zinc low remote data | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Fukutin-related protein, MAGNESIUM ION, ... | | Authors: | Kuwabara, N. | | Deposit date: | 2019-11-03 | | Release date: | 2020-01-15 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | Crystal structures of fukutin-related protein (FKRP), a ribitol-phosphate transferase related to muscular dystrophy.
Nat Commun, 11, 2020
|
|
6L7S
 
 | | Crystal structure of FKRP in complex with Mg ion, Zinc peak data | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Fukutin-related protein, MAGNESIUM ION, ... | | Authors: | Kuwabara, N. | | Deposit date: | 2019-11-03 | | Release date: | 2020-01-15 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | Crystal structures of fukutin-related protein (FKRP), a ribitol-phosphate transferase related to muscular dystrophy.
Nat Commun, 11, 2020
|
|
1GC3
 
 | | THERMUS THERMOPHILUS ASPARTATE AMINOTRANSFERASE TETRA MUTANT 2 COMPLEXED WITH TRYPTOPHAN | | Descriptor: | ASPARTATE AMINOTRANSFERASE, PYRIDOXAL-5'-PHOSPHATE, TRYPTOPHAN | | Authors: | Ura, H, Nakai, T, Hirotsu, K, Kuramitsu, S. | | Deposit date: | 2000-07-18 | | Release date: | 2001-09-05 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Substrate recognition mechanism of thermophilic dual-substrate enzyme.
J.Biochem., 130, 2001
|
|
1GC4
 
 | | THERMUS THERMOPHILUS ASPARTATE AMINOTRANSFERASE TETRA MUTANT 2 COMPLEXED WITH ASPARTATE | | Descriptor: | ASPARTATE AMINOTRANSFERASE, ASPARTIC ACID, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Ura, H, Nakai, T, Hirotsu, K, Kuramitsu, S. | | Deposit date: | 2000-07-19 | | Release date: | 2001-09-05 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Substrate recognition mechanism of thermophilic dual-substrate enzyme.
J.Biochem., 130, 2001
|
|
1GCK
 
 | | THERMUS THERMOPHILUS ASPARTATE AMINOTRANSFERASE DOUBLE MUTANT 1 COMPLEXED WITH ASPARTATE | | Descriptor: | ASPARTATE AMINOTRANSFERASE, ASPARTIC ACID, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Ura, H, Nakai, T, Hirotsu, K, Kuramitsu, S. | | Deposit date: | 2000-08-04 | | Release date: | 2001-11-14 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Substrate recognition mechanism of thermophilic dual-substrate enzyme.
J.Biochem., 130, 2001
|
|
4XKH
 
 | | CRYSTAL STRUCTURE OF THE AIRAPL TANDEM UIMS IN COMPLEX WITH A LYS48-LINKED TRI-UBIQUITIN | | Descriptor: | AN1-type zinc finger protein 2B, Polyubiquitin-C | | Authors: | Rahighi, S, Kawasaki, M, Stanhill, A, Wakatsuki, S. | | Deposit date: | 2015-01-11 | | Release date: | 2016-02-17 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Selective Binding of AIRAPL Tandem UIMs to Lys48-Linked Tri-Ubiquitin Chains.
Structure, 24, 2016
|
|
8ABT
 
 | | Crystal structure of NaLdpA in complex with the product analog Resveratrol | | Descriptor: | RESVERATROL, SULFATE ION, SnoaL-like domain-containing protein | | Authors: | Zahn, M, Kuatsjah, E, Beckham, G.T, McGeehan, J.E. | | Deposit date: | 2022-07-04 | | Release date: | 2023-02-01 | | Method: | X-RAY DIFFRACTION (1.39 Å) | | Cite: | Biochemical and structural characterization of a sphingomonad diarylpropane lyase for cofactorless deformylation.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8ABU
 
 | | Crystal structure of NaLdpA mutant H97Q in complex with erythro-DGPD | | Descriptor: | (1S,2R)-1,2-bis(3-methoxy-4-oxidanyl-phenyl)propane-1,3-diol, SnoaL-like domain-containing protein | | Authors: | Zahn, M, Kuatsjah, E, Beckham, G.T, McGeehan, J.E. | | Deposit date: | 2022-07-04 | | Release date: | 2023-02-01 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.661 Å) | | Cite: | Biochemical and structural characterization of a sphingomonad diarylpropane lyase for cofactorless deformylation.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8ABV
 
 | | Crystal structure of SpLdpA in complex with erythro-DGPD | | Descriptor: | (1R,2S)-1,2-bis(3-methoxy-4-oxidanyl-phenyl)propane-1,3-diol, (1S,2R)-1,2-bis(3-methoxy-4-oxidanyl-phenyl)propane-1,3-diol, GLYCEROL, ... | | Authors: | Zahn, M, Kuatsjah, E, Beckham, G.T, McGeehan, J.E. | | Deposit date: | 2022-07-04 | | Release date: | 2023-02-01 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.683 Å) | | Cite: | Biochemical and structural characterization of a sphingomonad diarylpropane lyase for cofactorless deformylation.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8ABW
 
 | | Crystal structure of SpLdpA in complex with threo-DGPD | | Descriptor: | (1R,2R)-1,2-bis(3-methoxy-4-oxidanyl-phenyl)propane-1,3-diol, (1S,2S)-1,2-bis(3-methoxy-4-oxidanyl-phenyl)propane-1,3-diol, SULFATE ION, ... | | Authors: | Zahn, M, Kuatsjah, E, Beckham, G.T, McGeehan, J.E. | | Deposit date: | 2022-07-04 | | Release date: | 2023-02-01 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Biochemical and structural characterization of a sphingomonad diarylpropane lyase for cofactorless deformylation.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
4WAA
 
 | | Crystal structure of Nix LIR-fused human LC3B_2-119 | | Descriptor: | Microtubule-associated proteins 1A/1B light chain 3B | | Authors: | Suzuki, H, Ravichandran, A.C, Dobson, R.C.J, Novak, I, Wakatsuki, S. | | Deposit date: | 2014-08-29 | | Release date: | 2015-09-09 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Phosphorylation of the mitochondrial autophagy receptor Nix enhances its interaction with LC3 proteins.
Sci Rep, 7, 2017
|
|
5XFE
 
 | | Luciferin-regenerating enzyme solved by SAD using XFEL (refined against 11,000 patterns) | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Luciferin regenerating enzyme, MAGNESIUM ION, ... | | Authors: | Yamashita, K, Pan, D, Okuda, T, Murai, T, Kodan, A, Yamaguchi, T, Gomi, K, Kajiyama, N, Kato, H, Ago, H, Yamamoto, M, Nakatsu, T. | | Deposit date: | 2017-04-10 | | Release date: | 2017-08-30 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Experimental phase determination with selenomethionine or mercury-derivatization in serial femtosecond crystallography
IUCrJ, 4, 2017
|
|
2LUE
 
 | | LC3B OPTN-LIR Ptot complex structure | | Descriptor: | Microtubule-associated proteins 1A/1B light chain 3B, Optineurin | | Authors: | Rogov, V.V, Rozenknop, A, Loehr, F, Guentert, P, Doetsch, V. | | Deposit date: | 2012-06-13 | | Release date: | 2013-07-17 | | Last modified: | 2022-08-24 | | Method: | SOLUTION NMR | | Cite: | Structural basis for phosphorylation-triggered autophagic clearance of Salmonella.
Biochem.J., 454, 2013
|
|
4G01
 
 | | ARA7-GDP-Ca2+/VPS9a | | Descriptor: | CALCIUM ION, GUANOSINE-5'-DIPHOSPHATE, Ras-related protein RABF2b, ... | | Authors: | Ihara, K. | | Deposit date: | 2012-07-09 | | Release date: | 2013-02-27 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Direct metal recognition by guanine nucleotide-exchange factor in the initial step of the exchange reaction
Acta Crystallogr.,Sect.D, 69, 2013
|
|
2A70
 
 | | Crystal structure of Emp47p carbohydrate recognition domain (CRD), monoclinic crystal form 2 | | Descriptor: | 1,2-ETHANEDIOL, Emp47p | | Authors: | Satoh, T, Sato, K, Kanoh, A, Yamashita, K, Katoh, R, Nakano, A, Wakatsuki, S. | | Deposit date: | 2005-07-04 | | Release date: | 2006-01-31 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Structures of the carbohydrate recognition domain of Ca2+-independent cargo receptors Emp46p and Emp47p.
J.Biol.Chem., 281, 2006
|
|
