4YNW
 
 | |
4ZOX
 
 | |
8PV8
 
 | | Chaetomium thermophilum pre-60S State 4 - post-5S rotation with Rix1 complex without Foot - composite structure | | Descriptor: | 26S rRNA, 5.8S rRNA, 5S rRNA, ... | | Authors: | Thoms, M, Cheng, J, Denk, T, Berninghausen, O, Beckmann, R. | | Deposit date: | 2023-07-17 | | Release date: | 2024-01-10 | | Last modified: | 2024-01-17 | | Method: | ELECTRON MICROSCOPY (2.91 Å) | | Cite: | Structural insights into coordinating 5S RNP rotation with ITS2 pre-RNA processing during ribosome formation.
Embo Rep., 24, 2023
|
|
8PV2
 
 | | Chaetomium thermophilum pre-60S State 10 - pre-5S rotation with Ytm1-Erb1 | | Descriptor: | 26S rRNA, 5.8S rRNA, 5S rRNA, ... | | Authors: | Thoms, M, Cheng, J, Denk, T, Berninghausen, O, Beckmann, R. | | Deposit date: | 2023-07-17 | | Release date: | 2023-11-15 | | Last modified: | 2023-12-20 | | Method: | ELECTRON MICROSCOPY (2.63 Å) | | Cite: | Structural insights into coordinating 5S RNP rotation with ITS2 pre-RNA processing during ribosome formation.
Embo Rep., 24, 2023
|
|
8PV4
 
 | | Chaetomium thermophilum pre-60S State 2 - pre-5S rotation with Rix1 complex - composite structure | | Descriptor: | 26S rRNA, 5.8S rRNA, 5S rRNA, ... | | Authors: | Thoms, M, Cheng, J, Denk, T, Berninghausen, O, Beckmann, R. | | Deposit date: | 2023-07-17 | | Release date: | 2023-11-15 | | Last modified: | 2023-12-20 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural insights into coordinating 5S RNP rotation with ITS2 pre-RNA processing during ribosome formation.
Embo Rep., 24, 2023
|
|
8PV3
 
 | | Chaetomium thermophilum pre-60S State 9 - pre-5S rotation - immature H68/H69 - composite structure | | Descriptor: | 26S rRNA, 5.8S rRNA, 5S rRNA, ... | | Authors: | Thoms, M, Cheng, J, Denk, T, Berninghausen, O, Beckmann, R. | | Deposit date: | 2023-07-17 | | Release date: | 2023-11-15 | | Last modified: | 2023-12-20 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structural insights into coordinating 5S RNP rotation with ITS2 pre-RNA processing during ribosome formation.
Embo Rep., 24, 2023
|
|
8PTW
 
 | | Chaetomium thermophilum Rix1-complex | | Descriptor: | Pre-rRNA-processing protein IPI3, Pre-rRNA-processing protein RIX1 | | Authors: | Thoms, M, Cheng, J, Denk, T, Berninghausen, O, Beckmann, R. | | Deposit date: | 2023-07-15 | | Release date: | 2023-11-15 | | Last modified: | 2023-12-20 | | Method: | ELECTRON MICROSCOPY (2.91 Å) | | Cite: | Structural insights into coordinating 5S RNP rotation with ITS2 pre-RNA processing during ribosome formation.
Embo Rep., 24, 2023
|
|
8PV6
 
 | | Chaetomium thermophilum pre-60S State 3 - post-5S rotation with Rix1 complex with Foot - composite structure | | Descriptor: | 26S rRNA, 5.8S rRNA, 5S rRNA, ... | | Authors: | Thoms, M, Cheng, J, Denk, T, Berninghausen, O, Beckmann, R. | | Deposit date: | 2023-07-17 | | Release date: | 2023-11-15 | | Last modified: | 2023-12-20 | | Method: | ELECTRON MICROSCOPY (2.94 Å) | | Cite: | Structural insights into coordinating 5S RNP rotation with ITS2 pre-RNA processing during ribosome formation.
Embo Rep., 24, 2023
|
|
8PUW
 
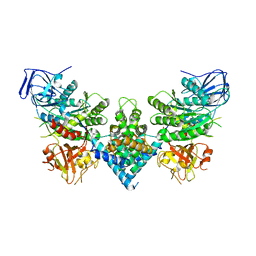 | | Chaetomium thermophilum Las1-Grc3-complex | | Descriptor: | Las1, Polynucleotide 5'-hydroxyl-kinase GRC3 | | Authors: | Thoms, M, Cheng, J, Denk, T, Berninghausen, O, Beckmann, R. | | Deposit date: | 2023-07-17 | | Release date: | 2023-11-15 | | Last modified: | 2023-12-20 | | Method: | ELECTRON MICROSCOPY (3.01 Å) | | Cite: | Structural insights into coordinating 5S RNP rotation with ITS2 pre-RNA processing during ribosome formation.
Embo Rep., 24, 2023
|
|
8PV1
 
 | | Chaetomium thermophilum pre-60S State 6 - pre-5S rotation - L1 intermediate - composite structure | | Descriptor: | 26S rRNA, 5.8S rRNA, 5S rRNA, ... | | Authors: | Thoms, M, Cheng, J, Denk, T, Berninghausen, O, Beckmann, R. | | Deposit date: | 2023-07-17 | | Release date: | 2023-11-15 | | Last modified: | 2023-12-20 | | Method: | ELECTRON MICROSCOPY (2.56 Å) | | Cite: | Structural insights into coordinating 5S RNP rotation with ITS2 pre-RNA processing during ribosome formation.
Embo Rep., 24, 2023
|
|
8PV5
 
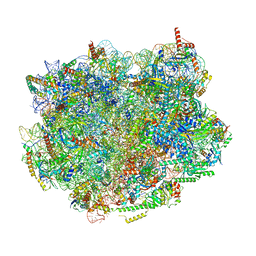 | | Chaetomium thermophilum pre-60S State 8 - pre-5S rotation without Foot - composite structure | | Descriptor: | 26S rRNA, 5.8S rRNA, 5S rRNA, ... | | Authors: | Thoms, M, Cheng, J, Denk, T, Berninghausen, O, Beckmann, R. | | Deposit date: | 2023-07-17 | | Release date: | 2023-11-15 | | Last modified: | 2023-12-20 | | Method: | ELECTRON MICROSCOPY (2.86 Å) | | Cite: | Structural insights into coordinating 5S RNP rotation with ITS2 pre-RNA processing during ribosome formation.
Embo Rep., 24, 2023
|
|
8PV7
 
 | | Chaetomium thermophilum pre-60S State 1 - pre-5S rotation (Arx1/Nog2 state) - Composite structure | | Descriptor: | 26S rRNA, 5.8S rRNA, 5S rRNA, ... | | Authors: | Thoms, M, Cheng, J, Denk, T, Berninghausen, O, Beckmann, R. | | Deposit date: | 2023-07-17 | | Release date: | 2023-11-15 | | Last modified: | 2023-12-20 | | Method: | ELECTRON MICROSCOPY (2.12 Å) | | Cite: | Structural insights into coordinating 5S RNP rotation with ITS2 pre-RNA processing during ribosome formation.
Embo Rep., 24, 2023
|
|
8PVL
 
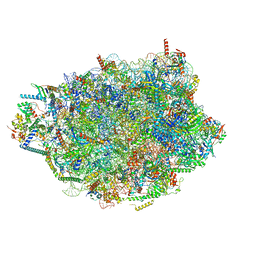 | | Chaetomium thermophilum pre-60S State 7 - pre-5S rotation lacking Utp30/ITS2 - composite structure | | Descriptor: | 26S rRNA, 5.8S rRNA, 5S rRNA, ... | | Authors: | Thoms, M, Cheng, J, Denk, T, Berninghausen, O, Beckmann, R. | | Deposit date: | 2023-07-17 | | Release date: | 2023-12-06 | | Last modified: | 2023-12-20 | | Method: | ELECTRON MICROSCOPY (2.19 Å) | | Cite: | Structural insights into coordinating 5S RNP rotation with ITS2 pre-RNA processing during ribosome formation.
Embo Rep., 24, 2023
|
|
8PVK
 
 | | Chaetomium thermophilum pre-60S State 5 - pre-5S rotation - L1 inward - composite structure | | Descriptor: | 26S rRNA, 5.8S rRNA, 5S rRNA, ... | | Authors: | Thoms, M, Cheng, J, Denk, T, Berninghausen, O, Beckmann, R. | | Deposit date: | 2023-07-17 | | Release date: | 2023-12-06 | | Last modified: | 2023-12-20 | | Method: | ELECTRON MICROSCOPY (2.55 Å) | | Cite: | Structural insights into coordinating 5S RNP rotation with ITS2 pre-RNA processing during ribosome formation.
Embo Rep., 24, 2023
|
|
4GMO
 
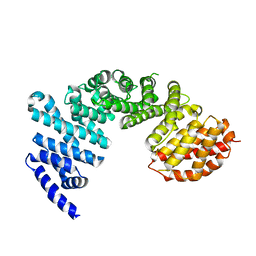 | | Crystal structure of Syo1 | | Descriptor: | Putative uncharacterized protein | | Authors: | Bange, G, Sinning, I. | | Deposit date: | 2012-08-16 | | Release date: | 2012-10-31 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Synchronizing nuclear import of ribosomal proteins with ribosome assembly.
Science, 338, 2012
|
|
4GMQ
 
 | | Ribosome-binding domain of Zuo1 | | Descriptor: | Putative ribosome associated protein | | Authors: | Kopp, J, Bange, G, Sinning, I. | | Deposit date: | 2012-08-16 | | Release date: | 2012-12-05 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structural characterization of a eukaryotic chaperone-the ribosome-associated complex.
Nat.Struct.Mol.Biol., 20, 2013
|
|
4GNI
 
 | | Structure of the Ssz1 ATPase bound to ATP and Magnesium | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, Putative heat shock protein | | Authors: | Bange, G, Sinning, I. | | Deposit date: | 2012-08-17 | | Release date: | 2012-12-05 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.796 Å) | | Cite: | Structural characterization of a eukaryotic chaperone-the ribosome-associated complex.
Nat.Struct.Mol.Biol., 20, 2013
|
|
6EMF
 
 | |
6EN7
 
 | |
6EMG
 
 | |
4IPA
 
 | | Structure of a thermophilic Arx1 | | Descriptor: | Putative curved DNA-binding protein, SULFATE ION | | Authors: | Bange, G, Sinning, I. | | Deposit date: | 2013-01-09 | | Release date: | 2013-01-30 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Consistent mutational paths predict eukaryotic thermostability.
BMC Evol Biol, 13, 2013
|
|
4GMN
 
 | | Structural basis of Rpl5 recognition by Syo1 | | Descriptor: | 60S ribosomal protein l5-like protein, Putative uncharacterized protein | | Authors: | Bange, G, Sinning, I. | | Deposit date: | 2012-08-16 | | Release date: | 2012-10-31 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Synchronizing nuclear import of ribosomal proteins with ribosome assembly.
Science, 338, 2012
|
|
4JQ5
 
 | |
4JNV
 
 | | Crystal structure of the human Nup57CCS3* coiled-coil segment, space group C2 | | Descriptor: | Nucleoporin p54 | | Authors: | Stuwe, T, Bley, C.J, Mayo, D.J, Hoelz, A. | | Deposit date: | 2013-03-15 | | Release date: | 2014-09-17 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Architecture of the fungal nuclear pore inner ring complex.
Science, 350, 2015
|
|
4JNU
 
 | | Crystal structure of the human Nup57CCS3* coiled-coil segment, space group P21 | | Descriptor: | Nucleoporin p54 | | Authors: | Stuwe, T, Bley, C.J, Mayo, D.J, Hoelz, A. | | Deposit date: | 2013-03-15 | | Release date: | 2014-09-17 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.445 Å) | | Cite: | Architecture of the fungal nuclear pore inner ring complex.
Science, 350, 2015
|
|
