7WKP
 
 | | ICP1 Csy4 | | Descriptor: | 3' stem loop crRNA, Csy4 | | Authors: | Zhang, M, Peng, R. | | Deposit date: | 2022-01-10 | | Release date: | 2023-04-26 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Mechanistic insights into DNA binding and cleavage by a compact type I-F CRISPR-Cas system in bacteriophage.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
7WWV
 
 | | DNA bound-ICP1 Csy complex | | Descriptor: | Csy1, Csy2, Csy3, ... | | Authors: | Zhang, M, Peng, R. | | Deposit date: | 2022-02-14 | | Release date: | 2023-04-26 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Mechanistic insights into DNA binding and cleavage by a compact type I-F CRISPR-Cas system in bacteriophage.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
7WWU
 
 | | ICP1 Csy complex | | Descriptor: | Csy1, Csy2, Csy3, ... | | Authors: | Zhang, M, Peng, R. | | Deposit date: | 2022-02-14 | | Release date: | 2023-04-26 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Mechanistic insights into DNA binding and cleavage by a compact type I-F CRISPR-Cas system in bacteriophage.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
7WKO
 
 | | ICP1 Csy1-2 complex | | Descriptor: | Csy1, Csy2 | | Authors: | Zhang, M, Peng, R. | | Deposit date: | 2022-01-10 | | Release date: | 2023-04-26 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Mechanistic insights into DNA binding and cleavage by a compact type I-F CRISPR-Cas system in bacteriophage.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
7Y82
 
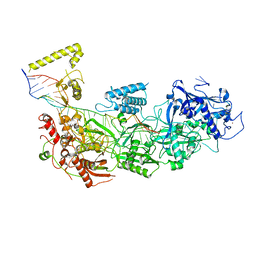 | | CryoEM structure of type III-E CRISPR Craspase gRAMP-crRNA complex bound to self RNA target | | Descriptor: | MAGNESIUM ION, RAMP superfamily protein, Self RNA target, ... | | Authors: | Zhang, J.T, Cui, N, Huang, H.D, Jia, N. | | Deposit date: | 2022-06-22 | | Release date: | 2022-12-14 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (2.83 Å) | | Cite: | Structural basis for the non-self RNA-activated protease activity of the type III-E CRISPR nuclease-protease Craspase.
Nat Commun, 13, 2022
|
|
7Y83
 
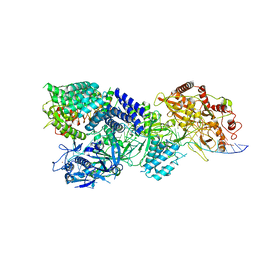 | | CryoEM structure of type III-E CRISPR Craspase gRAMP-crRNA in complex with TPR-CHAT protease bound to non-self RNA target | | Descriptor: | CHAT domain protein, MAGNESIUM ION, RAMP superfamily protein, ... | | Authors: | Zhang, J.T, Cui, N, Huang, H.D, Jia, N. | | Deposit date: | 2022-06-22 | | Release date: | 2022-12-14 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (2.93 Å) | | Cite: | Structural basis for the non-self RNA-activated protease activity of the type III-E CRISPR nuclease-protease Craspase.
Nat Commun, 13, 2022
|
|
7Y81
 
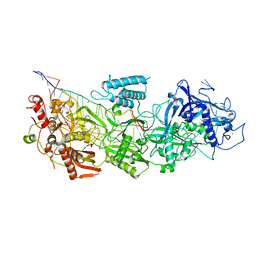 | | CryoEM structure of type III-E CRISPR Craspase gRAMP-crRNA complex bound to non-self RNA target | | Descriptor: | MAGNESIUM ION, Non-self RNA target, RAMP superfamily protein, ... | | Authors: | Zhang, J.T, Cui, N, Huang, H.D, Jia, N. | | Deposit date: | 2022-06-22 | | Release date: | 2022-12-14 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (2.54 Å) | | Cite: | Structural basis for the non-self RNA-activated protease activity of the type III-E CRISPR nuclease-protease Craspase.
Nat Commun, 13, 2022
|
|
7Y85
 
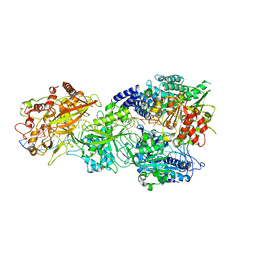 | | CryoEM structure of type III-E CRISPR Craspase gRAMP-crRNA in complex with TPR-CHAT protease bound to self RNA target | | Descriptor: | CHAT domain protein, MAGNESIUM ION, RAMP superfamily protein, ... | | Authors: | Zhang, J.T, Cui, N, Huang, H.D, Jia, N. | | Deposit date: | 2022-06-22 | | Release date: | 2022-12-14 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (2.73 Å) | | Cite: | Structural basis for the non-self RNA-activated protease activity of the type III-E CRISPR nuclease-protease Craspase.
Nat Commun, 13, 2022
|
|
7Y80
 
 | | CryoEM structure of type III-E CRISPR Craspase gRAMP-crRNA binary complex | | Descriptor: | MAGNESIUM ION, RAMP superfamily protein, ZINC ION, ... | | Authors: | Zhang, J.T, Cui, N, Huang, H.D, Jia, N. | | Deposit date: | 2022-06-22 | | Release date: | 2022-12-14 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (2.71 Å) | | Cite: | Structural basis for the non-self RNA-activated protease activity of the type III-E CRISPR nuclease-protease Craspase.
Nat Commun, 13, 2022
|
|
7Y84
 
 | | CryoEM structure of type III-E CRISPR Craspase gRAMP-crRNA in complex with TPR-CHAT protease | | Descriptor: | CHAT domain protein, MAGNESIUM ION, RAMP superfamily protein, ... | | Authors: | Zhang, J.T, Cui, N, Huang, H.D, Jia, N. | | Deposit date: | 2022-06-22 | | Release date: | 2022-12-14 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (2.61 Å) | | Cite: | Structural basis for the non-self RNA-activated protease activity of the type III-E CRISPR nuclease-protease Craspase.
Nat Commun, 13, 2022
|
|
7XP0
 
 | |
5Y7I
 
 | | Structure of tilapia fish CLIC2 | | Descriptor: | chloride intracellular channel protein 2 | | Authors: | Swaminathan, K, Zeng, J. | | Deposit date: | 2017-08-17 | | Release date: | 2018-03-21 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Tilapia and human CLIC2 structures are highly conserved.
Biochem. Biophys. Res. Commun., 495, 2018
|
|
5Y0I
 
 | | Solution structure of arenicin-3 derivative N1 | | Descriptor: | NZ17074(N1) | | Authors: | Liu, X.H, Wang, J.H. | | Deposit date: | 2017-07-17 | | Release date: | 2017-07-26 | | Last modified: | 2024-10-23 | | Method: | SOLUTION NMR | | Cite: | Antibacterial and detoxifying activity of NZ17074 analogues with multi-layers of selective antimicrobial actions against Escherichia coli and Salmonella enteritidis
Sci Rep, 7, 2017
|
|
5YL2
 
 | | Crystal structure of T2R-TTL-Y28 complex | | Descriptor: | (E)-1-(5-methoxy-2,2-dimethyl-chromen-8-yl)-3-(4-methoxy-3-oxidanyl-phenyl)prop-2-en-1-one, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, ... | | Authors: | Yang, J.H, Yang, T, Wen, J.L, Chen, L.J. | | Deposit date: | 2017-10-16 | | Release date: | 2018-04-18 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | The compound millepachine and its derivatives inhibit tubulin polymerization by irreversibly binding to the colchicine-binding site in beta-tubulin.
J. Biol. Chem., 2018
|
|
7W69
 
 | | Crystal structure of a PSH1 mutant in complex with EDO | | Descriptor: | 1,2-ETHANEDIOL, PSH1 | | Authors: | Gao, J, Lara, P, Li, Z.S, Han, X, Wei, R, Liu, W.D. | | Deposit date: | 2021-12-01 | | Release date: | 2022-09-14 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Multiple Substrate Binding Mode-Guided Engineering of a Thermophilic PET Hydrolase.
Acs Catalysis, 12, 2022
|
|
7W66
 
 | | Crystal structure of a PSH1 mutant in complex with ligand | | Descriptor: | PSH1, bis(2-hydroxyethyl) benzene-1,4-dicarboxylate | | Authors: | Gao, J, Lara, P, Li, Z.S, Han, X, Wei, R, Liu, W.D. | | Deposit date: | 2021-12-01 | | Release date: | 2022-09-14 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Multiple Substrate Binding Mode-Guided Engineering of a Thermophilic PET Hydrolase.
Acs Catalysis, 12, 2022
|
|
7W6C
 
 | | Crystal structure of a PSH1 in complex with ligand J1K | | Descriptor: | 4-(2-hydroxyethylcarbamoyl)benzoic acid, PSH1 | | Authors: | Gao, J, Lara, P, Li, Z.S, Han, X, Wei, R, Liu, W.D. | | Deposit date: | 2021-12-01 | | Release date: | 2022-09-14 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Multiple Substrate Binding Mode-Guided Engineering of a Thermophilic PET Hydrolase.
Acs Catalysis, 12, 2022
|
|
7W6O
 
 | | Crystal structure of a PSH1 in complex with J1K | | Descriptor: | 4-(2-hydroxyethylcarbamoyl)benzoic acid, PSH1 | | Authors: | Gao, J, Lara, P, Li, Z.S, Han, X, Wei, R, Liu, W.D. | | Deposit date: | 2021-12-02 | | Release date: | 2022-09-14 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Multiple Substrate Binding Mode-Guided Engineering of a Thermophilic PET Hydrolase.
Acs Catalysis, 12, 2022
|
|
7W6Q
 
 | | Crystal structure of a PSH1 in complex with ligand J1K | | Descriptor: | 4-(2-hydroxyethylcarbamoyl)benzoic acid, PSH1 | | Authors: | Gao, J, Lara, P, Li, Z.S, Han, X, Wei, R, Liu, W.D. | | Deposit date: | 2021-12-02 | | Release date: | 2022-09-14 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Multiple Substrate Binding Mode-Guided Engineering of a Thermophilic PET Hydrolase.
Acs Catalysis, 12, 2022
|
|
5Y0H
 
 | | Solution structure of arenicin-3 derivative N6 | | Descriptor: | N6 | | Authors: | Liu, X.H, Wang, J.H. | | Deposit date: | 2017-07-17 | | Release date: | 2017-07-26 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | Antibacterial and detoxifying activity of NZ17074 analogues with multi-layers of selective antimicrobial actions against Escherichia coli and Salmonella enteritidis
Sci Rep, 7, 2017
|
|
5Y0J
 
 | | Solution structure of arenicin-3 derivative N2 | | Descriptor: | N2 | | Authors: | Liu, X.H, Wang, J.H. | | Deposit date: | 2017-07-17 | | Release date: | 2017-07-26 | | Last modified: | 2024-11-06 | | Method: | SOLUTION NMR | | Cite: | Antibacterial and detoxifying activity of NZ17074 analogues with multi-layers of selective antimicrobial actions against Escherichia coli and Salmonella enteritidis
Sci Rep, 7, 2017
|
|
7W16
 
 | | Complex structure of alginate lyase AlyV with M8 | | Descriptor: | GLYCEROL, alginate lyase, beta-D-mannopyranuronic acid-(1-4)-beta-D-mannopyranuronic acid | | Authors: | Liu, W.Z, Lyu, Q.Q, Li, Z.J, Zhang, K.K. | | Deposit date: | 2021-11-19 | | Release date: | 2022-08-17 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Determination of oligosaccharide product distributions of PL7 alginate lyases by their structural elements.
Commun Biol, 5, 2022
|
|
7W13
 
 | | Complex structure of alginate lyase PyAly with M8 | | Descriptor: | Alginate lyase, beta-D-mannopyranuronic acid-(1-4)-beta-D-mannopyranuronic acid-(1-4)-beta-D-mannopyranuronic acid | | Authors: | Liu, W.Z, Lyu, Q.Q, Li, Z.J, Zhang, K.K. | | Deposit date: | 2021-11-19 | | Release date: | 2022-08-17 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Determination of oligosaccharide product distributions of PL7 alginate lyases by their structural elements.
Commun Biol, 5, 2022
|
|
7XRT
 
 | | Bacteroides thetaiotaomicron ferulic acid esterase (BT_4077) | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CALCIUM ION, Ferulic acid esterase | | Authors: | Du, G.M, Wang, Y.L, Xin, F.J. | | Deposit date: | 2022-05-11 | | Release date: | 2023-11-22 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.007 Å) | | Cite: | Structural insights into the oligomeric effects on catalytic activity of a decameric feruloyl esterase and its application in ferulic acid production.
Int.J.Biol.Macromol., 253, 2023
|
|
6A3N
 
 | | Crystal structure of the PDE9 catalytic domain in complex with inhibitor 2 | | Descriptor: | 1-cyclopentyl-6-({(2R)-1-[(3S)-3-fluoropyrrolidin-1-yl]-1-oxopropan-2-yl}amino)-1,5-dihydro-4H-pyrazolo[3,4-d]pyrimidin-4-one, High affinity cGMP-specific 3',5'-cyclic phosphodiesterase 9A, MAGNESIUM ION, ... | | Authors: | Wu, Y.N, Zhou, Q, Chen, Y.P, Luo, H.B. | | Deposit date: | 2018-06-15 | | Release date: | 2019-04-10 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Discovery of Potent, Selective, and Orally Bioavailable Inhibitors against Phosphodiesterase-9, a Novel Target for the Treatment of Vascular Dementia.
J. Med. Chem., 62, 2019
|
|
