4CS1
 
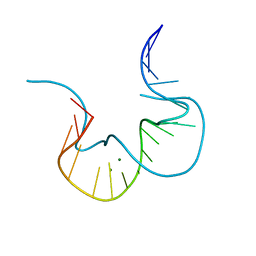 | |
6G42
 
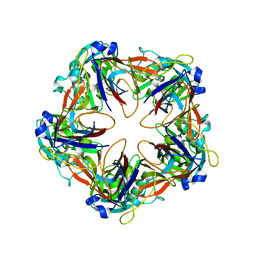 | | Crystal structure of mavirus penton protein | | Descriptor: | Minor capsid protein | | Authors: | Born, D, Reuter, L, Meinhart, A, Reinstein, J. | | Deposit date: | 2018-03-26 | | Release date: | 2018-07-04 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Capsid protein structure, self-assembly, and processing reveal morphogenesis of the marine virophage mavirus.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
4COD
 
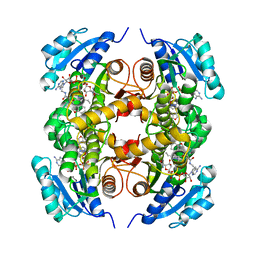 | | Encoded library technology as a source of hits for the discovery and lead optimization of a potent and selective class of bactericidal direct inhibitors of Mycobacterium tuberculosis InhA | | Descriptor: | ENOYL-[ACYL-CARRIER-PROTEIN] REDUCTASE [NADH], N-((3R,5S)-1-(benzofuran-3-carbonyl)-5-(ethylcarbamoyl)pyrrolidin-3-yl)-3-ethyl-1-methyl-1H-pyrazole-5-carboxamide, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Encinas, L, OKeefe, H, Neu, M, Convery, M.A, McDowell, W, Mendoza-Losana, A, Pages, L.B, Castro-Pichel, J, Evindar, G. | | Deposit date: | 2014-01-28 | | Release date: | 2014-02-12 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Encoded Library Technology as a Source of Hits for the Discovery and Lead Optimization of a Potent and Selective Class of Bactericidal Direct Inhibitors of Mycobacterium Tuberculosis Inha.
J.Med.Chem., 57, 2014
|
|
4CP3
 
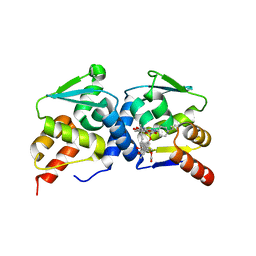 | | The structure of BCL6 BTB (POZ) domain in complex with the ansamycin antibiotic rifabutin. | | Descriptor: | B-CELL LYMPHOMA 6 PROTEIN, RIFABUTIN | | Authors: | Evans, S.E, Fairall, L, Goult, B.T, Jamieson, A.G, Ferrigno, P.K, Ford, R, Wagner, S.D, Schwabe, J.W.R. | | Deposit date: | 2014-01-31 | | Release date: | 2014-03-19 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The Ansamycin Antibiotic, Rifamycin Sv, Inhibits Bcl6 Transcriptional Repression and Forms a Complex with the Bcl6-Btb/Poz Domain.
Plos One, 9, 2014
|
|
6G6O
 
 | | Crystal structure of the computationally designed Ika8 protein: crystal packing No.1 in P63 | | Descriptor: | GLYCEROL, Ika8 | | Authors: | Noguchi, H, Addy, C, Simoncini, D, Van Meervelt, L, Schiex, T, Zhang, K.Y.J, Tame, J.R.H, Voet, A.R.D. | | Deposit date: | 2018-04-01 | | Release date: | 2018-11-28 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Computational design of symmetrical eight-bladed beta-propeller proteins.
IUCrJ, 6, 2019
|
|
5OBW
 
 | | Mycoplasma genitalium DnaK-NBD in complex with ATP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Chaperone protein DnaK, GLYCEROL, ... | | Authors: | Adell, M, Calisto, B, Fita, I, Martinelli, L. | | Deposit date: | 2017-06-29 | | Release date: | 2018-05-09 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | The nucleotide-bound/substrate-bound conformation of the Mycoplasma genitalium DnaK chaperone.
Protein Sci., 27, 2018
|
|
5OCH
 
 | | The crystal structure of human ABCB8 in an outward-facing state | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATP-binding cassette sub-family B member 8, mitochondrial, ... | | Authors: | Faust, B, Pike, A.C.W, Shintre, C.A, Quiqley, A.M, Chu, A, Barr, A, Shrestha, L, Mukhopadhyay, S, Borkowska, O, Chalk, R, Burgess-Brown, N.A, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Carpenter, E.P, Structural Genomics Consortium (SGC) | | Deposit date: | 2017-06-30 | | Release date: | 2018-07-25 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | The crystal structure of human ABCB8 in an outward-facing state
To Be Published
|
|
4CWA
 
 | | Structure of Plasmodium Falciparum Spermidine Synthase in Complex with 1H-Benzimidazole-2-pentanamine | | Descriptor: | 2-(2-{2-[2-(2-METHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHANOL, 5-(1H-benzimidazol-2-yl)pentan-1-amine, SPERMIDINE SYNTHASE | | Authors: | Sprenger, J, Halander, J.C, Svensson, B, Al-Karadaghi, S, Persson, L. | | Deposit date: | 2014-04-01 | | Release date: | 2014-10-08 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Three-Dimensional Structures of Plasmodium Falciparum Spermidine Synthase with Bound Inhibitors Suggest New Strategies for Drug Design.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
8S8I
 
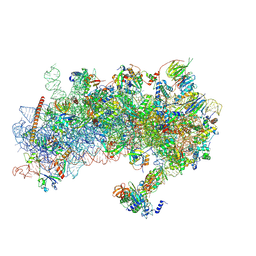 | | Structure of a yeast 48S-AUC preinitiation complex in closed conformation (model py48S-AUC-eIF1) | | Descriptor: | 18S ribosomal RNA, 40S ribosomal protein L41-A, 40S ribosomal protein S12, ... | | Authors: | Villamayor-Belinchon, L, Sharma, P, Llacer, J.L, Hussain, T. | | Deposit date: | 2024-03-06 | | Release date: | 2024-09-11 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Structural basis of AUC codon discrimination during translation initiation in yeast.
Nucleic Acids Res., 52, 2024
|
|
3UU5
 
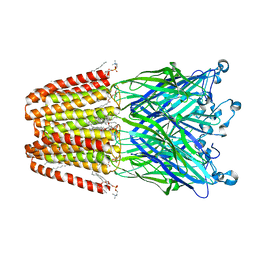 | | The GLIC pentameric Ligand-Gated Ion Channel Loop2-20' mutant reduced in solution | | Descriptor: | CHLORIDE ION, DIUNDECYL PHOSPHATIDYL CHOLINE, DODECYL-BETA-D-MALTOSIDE, ... | | Authors: | Sauguet, L, Nury, H, Corringer, P.J, Delarue, M. | | Deposit date: | 2011-11-28 | | Release date: | 2012-05-16 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | A locally closed conformation of a bacterial pentameric proton-gated ion channel.
Nat.Struct.Mol.Biol., 19, 2012
|
|
4CYE
 
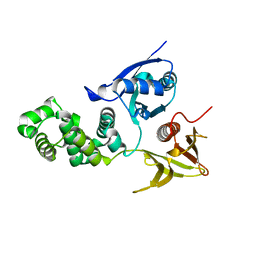 | | Crystal structure of avian FAK FERM domain FAK31-405 at 3.2A | | Descriptor: | FOCAL ADHESION KINASE 1 | | Authors: | Goni, G.M, Epifano, C, Boskovic, J, Camacho-Artacho, M, Zhou, J, Martin, M.T, Eck, M.J, Kremer, L, Graeter, F, Gervasio, F.L, Perez-Moreno, M, Lietha, D. | | Deposit date: | 2014-04-10 | | Release date: | 2014-04-23 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Phosphatidylinositol 4,5-Bisphosphate Triggers Activation of Focal Adhesion Kinase by Inducing Clustering and Conformational Changes.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
3V0S
 
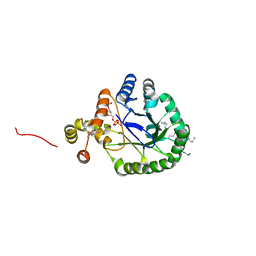 | | Crystal Structure of Perakine Reductase, Founder Member of a Novel AKR Subfamily with Unique Conformational Changes during NADPH Binding | | Descriptor: | 2'-MONOPHOSPHOADENOSINE-5'-DIPHOSPHATE, Perakine reductase | | Authors: | Sun, L, Chen, Y, Rajendran, C, Panjikar, S, Mueller, U, Wang, M, Rosenthal, C, Mindnich, R, Penning, T.M, Stoeckigt, J. | | Deposit date: | 2011-12-08 | | Release date: | 2012-02-22 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.773 Å) | | Cite: | Crystal structure of perakine reductase, founding member of a novel aldo-keto reductase (AKR) subfamily that undergoes unique conformational changes during NADPH binding.
J.Biol.Chem., 287, 2012
|
|
8R3E
 
 | | Huntingtin, 1-17, MBP-N | | Descriptor: | Maltodextrin-binding protein, ZINC ION | | Authors: | Steinbacher, S, Toledo-Sherman, L, Dominguez, C. | | Deposit date: | 2023-11-08 | | Release date: | 2024-09-18 | | Method: | X-RAY DIFFRACTION (2.91 Å) | | Cite: | Post-translational modifications in the first 17 amino acids of huntingtin influence self-association and interaction with membranes
To Be Published
|
|
5OBY
 
 | | Mycoplasma genitalium DnaK-NBD in complex with AMP-PNP | | Descriptor: | Chaperone protein DnaK, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Adell, M, Calisto, B, Fita, I, Martinelli, L. | | Deposit date: | 2017-06-29 | | Release date: | 2018-05-09 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | The nucleotide-bound/substrate-bound conformation of the Mycoplasma genitalium DnaK chaperone.
Protein Sci., 27, 2018
|
|
5XKD
 
 | | Crystal structure of dibenzothiophene sulfone monooxygenase BdsA in complex with FMN at 2.4 angstrom | | Descriptor: | Dibenzothiophene desulfurization enzyme A, FLAVIN MONONUCLEOTIDE | | Authors: | Gu, L, Su, T, Liu, S, Su, J. | | Deposit date: | 2017-05-07 | | Release date: | 2018-05-09 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.393 Å) | | Cite: | Structural and Biochemical Characterization of BdsA fromBacillus subtilisWU-S2B, a Key Enzyme in the "4S" Desulfurization Pathway.
Front Microbiol, 9, 2018
|
|
8S8D
 
 | | Structure of a yeast 48S-AUC preinitiation complex in closed conformation (model py48S-AUC-2) | | Descriptor: | 18S ribosomal RNA, 40S ribosomal protein L41-A, 40S ribosomal protein S12, ... | | Authors: | Villamayor-Belinchon, L, Sharma, P, Llacer, J.L, Hussain, T. | | Deposit date: | 2024-03-06 | | Release date: | 2024-09-11 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.45 Å) | | Cite: | Structural basis of AUC codon discrimination during translation initiation in yeast.
Nucleic Acids Res., 52, 2024
|
|
3UYI
 
 | | Crystal Structure of Perakine Reductase, Founder Member of a Novel AKR Subfamily with Unique Conformational Changes during NADPH Binding | | Descriptor: | Perakine reductase | | Authors: | Sun, L, Chen, Y, Rajendran, C, Panjikar, S, Mueller, U, Wang, M, Rosenthal, C, Mindnich, R, Penning, T.M, Stoeckigt, J. | | Deposit date: | 2011-12-06 | | Release date: | 2012-02-22 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.313 Å) | | Cite: | Crystal structure of perakine reductase, founding member of a novel aldo-keto reductase (AKR) subfamily that undergoes unique conformational changes during NADPH binding.
J.Biol.Chem., 287, 2012
|
|
8S8G
 
 | | Structure of a yeast 48S-AUC preinitiation complex in closed conformation (model py48S-AUC-2.1) | | Descriptor: | 18S ribosomal RNA, 40S ribosomal protein L41-A, 40S ribosomal protein S12, ... | | Authors: | Villamayor-Belinchon, L, Sharma, P, Llacer, J.L, Hussain, T. | | Deposit date: | 2024-03-06 | | Release date: | 2024-09-11 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structural basis of AUC codon discrimination during translation initiation in yeast.
Nucleic Acids Res., 52, 2024
|
|
8XLP
 
 | | Structure of inactive Photosystem II associated with CAC antenna from Rhodomonas Salina | | Descriptor: | (1~{R})-3,5,5-trimethyl-4-[(3~{E},5~{E},7~{E},9~{E},11~{E},13~{E},15~{E})-3,7,12,16-tetramethyl-18-[(4~{R})-2,6,6-trimethyl-4-oxidanyl-cyclohexen-1-yl]octadeca-3,5,7,9,11,13,15-heptaen-1,17-diynyl]cyclohex-3-en-1-ol, (1~{R})-3,5,5-trimethyl-4-[(3~{E},5~{E},7~{E},9~{E},11~{E},13~{E},15~{E},17~{E})-3,7,12,16-tetramethyl-18-(2,6,6-trimethylcyclohexen-1-yl)octadeca-3,5,7,9,11,13,15,17-octaen-1-ynyl]cyclohex-3-en-1-ol, 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, ... | | Authors: | Si, L, Li, M. | | Deposit date: | 2023-12-26 | | Release date: | 2024-07-31 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.57 Å) | | Cite: | Structural basis for the distinct core-antenna assembly of cryptophyte photosystem II.
Nat Commun, 15, 2024
|
|
1FZF
 
 | | CRYSTAL STRUCTURE OF FRAGMENT DOUBLE-D FROM HUMAN FIBRIN WITH THE PEPTIDE LIGAND GLY-HIS-ARG-PRO-AMIDE | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, FIBRINOGEN | | Authors: | Everse, S.J, Spraggon, G, Veerapandian, L, Doolittle, R.F. | | Deposit date: | 1998-12-28 | | Release date: | 1999-06-08 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Conformational changes in fragments D and double-D from human fibrin(ogen) upon binding the peptide ligand Gly-His-Arg-Pro-amide.
Biochemistry, 38, 1999
|
|
3V49
 
 | | Structure of ar lbd with activator peptide and sarm inhibitor 1 | | Descriptor: | 4-[(4R)-4-(4-hydroxyphenyl)-3,4-dimethyl-2,5-dioxoimidazolidin-1-yl]-2-(trifluoromethyl)benzonitrile, Androgen receptor, activator peptide, ... | | Authors: | Nique, F, Hebbe, S, Peixoto, C, Annoot, D, Lefrancois, J.-M, Duval, E, Michoux, L, Triballeau, N, Lemoullec, J.-M, Mollat, P, Thauvin, M, Prange, T, Minet, D, Clement-Lacroix, P, Robin-Jagerschmidt, C, Fleury, D, Guedin, D, Deprez, P. | | Deposit date: | 2011-12-14 | | Release date: | 2012-09-05 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Discovery of diarylhydantoins as new selective androgen receptor modulators.
J.Med.Chem., 55, 2012
|
|
6FZ0
 
 | |
8S8H
 
 | | Structure of a yeast 48S-AUC preinitiation complex in closed conformation (model py48S-AUC-2.2) | | Descriptor: | 18S ribosomal RNA, 40S ribosomal protein L41-A, 40S ribosomal protein S12, ... | | Authors: | Villamayor-Belinchon, L, Sharma, P, Llacer, J.L, Hussain, T. | | Deposit date: | 2024-03-06 | | Release date: | 2024-09-11 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structural basis of AUC codon discrimination during translation initiation in yeast.
Nucleic Acids Res., 52, 2024
|
|
3V0E
 
 | | Crystal structure of Ciona intestinalis voltage sensor-containing phosphatase (Ci-VSP), residues 256-576(C363S) | | Descriptor: | CHLORIDE ION, GLYCEROL, SULFATE ION, ... | | Authors: | Liu, L, Kohout, S.C, Xu, Q, Muller, S, Kimberlin, C, Isacoff, E.Y, Minor, D.L. | | Deposit date: | 2011-12-07 | | Release date: | 2012-05-09 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | A glutamate switch controls voltage-sensitive phosphatase function.
Nat.Struct.Mol.Biol., 19, 2012
|
|
5XMK
 
 | | Cryo-EM structure of the ATP-bound Vps4 mutant-E233Q complex with Vta1 (masked) | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Vacuolar protein sorting-associated protein 4, Vacuolar protein sorting-associated protein VTA1 | | Authors: | Sun, S, Li, L, Yang, F, Wang, X, Fan, F, Li, X, Wang, H, Sui, S. | | Deposit date: | 2017-05-15 | | Release date: | 2017-08-09 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (4.18 Å) | | Cite: | Cryo-EM structures of the ATP-bound Vps4(E233Q) hexamer and its complex with Vta1 at near-atomic resolution
Nat Commun, 8, 2017
|
|
