8UWS
 
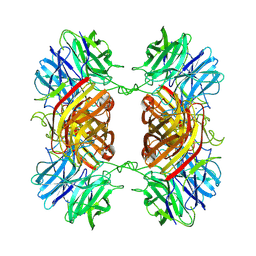 | | Cryo-EM structure of an Enterobacter GH43 Beta-Xylosidase: EcXyl43 | | Descriptor: | Beta-xylosidase, CALCIUM ION | | Authors: | Briganti, L, Godoy, A.S, Capetti, C.C.M, Portugal, R.V, Polikarpov, I. | | Deposit date: | 2023-11-08 | | Release date: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (2.65 Å) | | Cite: | Cryo-EM structure of an Enterobacter GH43 Beta-Xylosidase: EcXyl43
To Be Published
|
|
6AKK
 
 | | Crystal structure of the second Coiled-coil domain of SIKE1 | | Descriptor: | GLYCEROL, Suppressor of IKBKE 1 | | Authors: | Zhou, L, Chen, M, Zhou, Z.C. | | Deposit date: | 2018-09-02 | | Release date: | 2019-01-16 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Architecture, substructures, and dynamic assembly of STRIPAK complexes in Hippo signaling.
Cell Discov, 5, 2019
|
|
4CXM
 
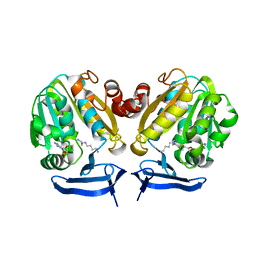 | | Crystal Structure of Plasmodium Falciparum Spermidine Synthase in Complex with METHYLTHIOADENOSIN AND SPERMIDINE after catalysis in crystal | | Descriptor: | 5'-DEOXY-5'-METHYLTHIOADENOSINE, SPERMIDINE, SPERMIDINE SYNTHASE | | Authors: | Sprenger, J, Svensson, B, Al-Karadaghi, S, Persson, L. | | Deposit date: | 2014-04-07 | | Release date: | 2014-10-08 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Three-Dimensional Structures of Plasmodium Falciparum Spermidine Synthase with Bound Inhibitors Suggest New Strategies for Drug Design.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
1NBW
 
 | | Glycerol dehydratase reactivase | | Descriptor: | CALCIUM ION, GLYCEROL DEHYDRATASE REACTIVASE ALPHA SUBUNIT, GLYCEROL DEHYDRATASE REACTIVASE BETA SUBUNIT | | Authors: | Liao, D.-I, Reiss, L, Turner Jr, I, Dotson, G. | | Deposit date: | 2002-12-04 | | Release date: | 2003-01-14 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of glycerol dehydratase reactivase: A new type of molecular chaperone
Structure, 11, 2003
|
|
8BLT
 
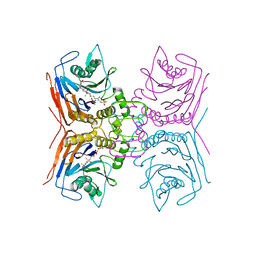 | | Structure of Lactobacillus salivarius (Ls) bile salt hydrolase(BSH) in complex with taurocholate (TCA) | | Descriptor: | Bile salt hydrolase, TAUROCHOLIC ACID | | Authors: | Karlov, D.S, Long, S.L, Zeng, X, Xu, F, Lal, K, Cao, L, Hayoun, K, Lin, J, Joyce, S.A, Tikhonova, I.G. | | Deposit date: | 2022-11-10 | | Release date: | 2023-03-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Characterization of the mechanism of bile salt hydrolase substrate specificity by experimental and computational analyses.
Structure, 31, 2023
|
|
8V7T
 
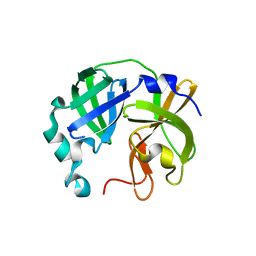 | |
4CXY
 
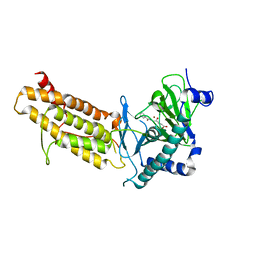 | | Crystal structure of human FTO in complex with acylhydrazine inhibitor 21 | | Descriptor: | (E)-4-(2-Nicotinoylhydrazinyl)-4-oxobut-2-enoic acid, ALPHA-KETOGLUTARATE-DEPENDENT DIOXYGENASE FTO, NICKEL (II) ION | | Authors: | Toh, D.W, Sun, L, Tan, J, Chen, Y, Lau, L.Z.M, Hong, W, Woon, E.C.Y, Gao, Y.G. | | Deposit date: | 2014-04-09 | | Release date: | 2014-10-01 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | A strategy based on nucleotide specificity leads to a subfamily-selective and cell-active inhibitor ofN6-methyladenosine demethylase FTO.
Chem Sci, 6, 2015
|
|
1NDI
 
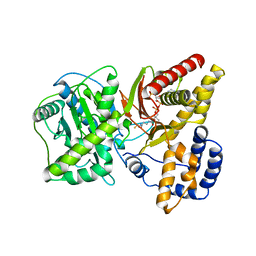 | | Carnitine Acetyltransferase in complex with CoA | | Descriptor: | COENZYME A, Carnitine Acetyltransferase | | Authors: | Jogl, G, Tong, L. | | Deposit date: | 2002-12-09 | | Release date: | 2003-01-28 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structure of Carnitine Acetyltransferase and Implications for the Catalytic Mechanism and Fatty Acid Transport
Cell(Cambridge,Mass.), 112, 2003
|
|
5NWQ
 
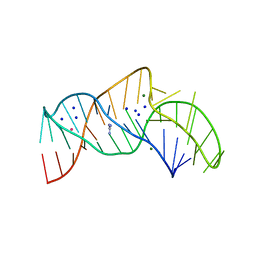 | | The structure of the thermobifida fusca guanidine III riboswitch with guanidine. | | Descriptor: | GUANIDINE, Guanidine III riboswitch, MAGNESIUM ION, ... | | Authors: | Huang, L, Wang, J, Lilley, D.M.J. | | Deposit date: | 2017-05-08 | | Release date: | 2017-10-18 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Structure of the Guanidine III Riboswitch.
Cell Chem Biol, 24, 2017
|
|
1EW6
 
 | |
8QZJ
 
 | |
5NWX
 
 | | Insight into the molecular recognition mechanism of the coactivator NCoA1 by STAT6 | | Descriptor: | Nuclear receptor coactivator 1, Signal transducer and activator of transcription 6 | | Authors: | Russo, L, Giller, K, Pfitzner, E, Griesinger, C, Becker, S. | | Deposit date: | 2017-05-08 | | Release date: | 2017-12-13 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Insight into the molecular recognition mechanism of the coactivator NCoA1 by STAT6.
Sci Rep, 7, 2017
|
|
6H1W
 
 | |
8QZI
 
 | | Crystal structure of PptT-ACP from Mycobacterium tuberculosis | | Descriptor: | 4'-phosphopantetheinyl transferase PptT, COENZYME A, ISOPROPYL ALCOHOL, ... | | Authors: | Gavalda, S, Mourey, L, Pedelacq, J.D. | | Deposit date: | 2023-10-27 | | Release date: | 2024-06-19 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Catalytic Cycle of Type II 4'-Phosphopantetheinyl Transferases
Acs Catalysis, 14, 2024
|
|
1NHU
 
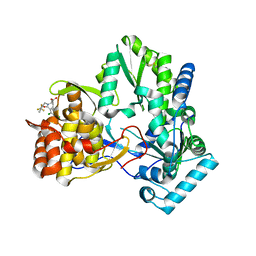 | | Hepatitis C virus RNA polymerase in complex with non-nucleoside analogue inhibitor | | Descriptor: | (2S)-2-[(2,4-DICHLORO-BENZOYL)-(3-TRIFLUOROMETHYL-BENZYL)-AMINO]-3-PHENYL-PROPIONIC ACID, HEPATITIS C VIRUS NS5B RNA-DEPENDENT RNA POLYMERASE | | Authors: | Wang, M, Ng, K.K.S, Cherney, M.M, Chan, L, Yannopoulos, C.G, Bedard, J, Morin, N, Nguyen-Ba, N, Alaoui-Ismaili, M.H, Bethell, R.C, James, M.N.G. | | Deposit date: | 2002-12-19 | | Release date: | 2003-03-18 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Non-Nucleoside Analogue Inhibitors Bind to an Allosteric Site on
HCV NS5B Polymerase: Crystal Structures and Mechanism of Inhibition
J.Biol.Chem., 278, 2003
|
|
5ZVT
 
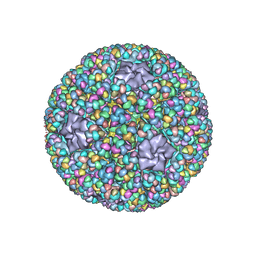 | | Structure of RNA polymerase complex and genome within a dsRNA virus provides insights into the mechanisms of transcription and assembly | | Descriptor: | C-terminus of outer capsid protein VP5, Core protein VP6, MYRISTIC ACID, ... | | Authors: | Liu, H, Fang, Q, Cheng, L. | | Deposit date: | 2018-05-12 | | Release date: | 2018-07-04 | | Last modified: | 2018-07-25 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structure of RNA polymerase complex and genome within a dsRNA virus provides insights into the mechanisms of transcription and assembly.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6H37
 
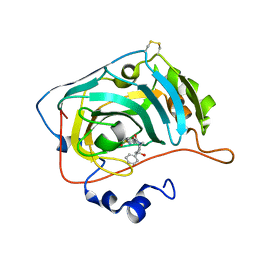 | | The crystal structure of human carbonic anhydrase VII in complex with 4-(4-phenyl)-4-hydroxy-1-piperidine-1-carbonyl)benzenesulfonamide | | Descriptor: | 4-(4-oxidanyl-4-phenyl-piperidin-1-yl)carbonylbenzenesulfonamide, Carbonic anhydrase 7, ZINC ION | | Authors: | Buemi, M.R, Di Fiore, A, De Luca, L, Ferro, S, Mancuso, F, Monti, S.M, Buonanno, M, Angeli, A, Russo, E, De Sarro, G, Supuran, C.T, De Simone, G, Gitto, R. | | Deposit date: | 2018-07-17 | | Release date: | 2018-12-19 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Exploring structural properties of potent human carbonic anhydrase inhibitors bearing a 4-(cycloalkylamino-1-carbonyl)benzenesulfonamide moiety.
Eur J Med Chem, 163, 2018
|
|
4CXX
 
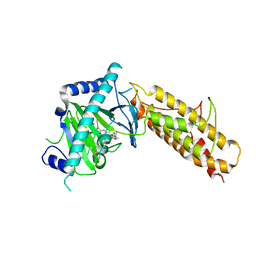 | | Crystal structure of human FTO in complex with acylhydrazine inhibitor 16 | | Descriptor: | (2E)-4-{N'-[4-(4-tert-Butyl-benzyl)-pyridine-3-carbonyl]-hydrazino}-4-oxo-but-2-enoic acid, ALPHA-KETOGLUTARATE-DEPENDENT DIOXYGENASE FTO, NICKEL (II) ION | | Authors: | Toh, D.W, Sun, L, Tan, J, Chen, Y, Lau, L.Z.M, Hong, W, Woon, E.C.Y, Gao, Y.G. | | Deposit date: | 2014-04-09 | | Release date: | 2014-10-01 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.76 Å) | | Cite: | A strategy based on nucleotide specificity leads to a subfamily-selective and cell-active inhibitor ofN6-methyladenosine demethylase FTO.
Chem Sci, 6, 2015
|
|
3V6J
 
 | | Replication of N2,3-Ethenoguanine by DNA Polymerases | | Descriptor: | 2'-DEOXYCYTIDINE-5'-TRIPHOSPHATE, DNA (5'-D(*GP*GP*GP*GP*AP*AP*GP*GP*AP*TP*TP*(DOC))-3'), DNA (5'-D(*TP*CP*AP*TP*(EFG)P*GP*AP*AP*TP*CP*CP*TP*TP*CP*CP*CP*C)-3'), ... | | Authors: | Zhao, L. | | Deposit date: | 2011-12-19 | | Release date: | 2012-04-25 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Replication of n(2) ,3-ethenoguanine by DNA polymerases.
Angew.Chem.Int.Ed.Engl., 51, 2012
|
|
1N9I
 
 | | structure of earth-grown oxidized myoglobin mutant YQR (ISS8A) | | Descriptor: | HYDROXIDE ION, Myoglobin, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Miele, A.E, Sciara, G, Federici, L, Vallone, B, Brunori, M. | | Deposit date: | 2002-11-25 | | Release date: | 2003-06-10 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Analysis of the effect of microgravity on protein crystal quality: the case of a myoglobin triple mutant.
Acta Crystallogr.,Sect.D, 59, 2003
|
|
4CRQ
 
 | | Crystal structure of the catalytic domain of the modular laminarinase ZgLamC mutant E142S | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, CALCIUM ION, ... | | Authors: | Labourel, A, Jam, M, Legentil, L, Sylla, B, Ficko-Blean, E, Hehemann, J.H, Ferrieres, V, Czjzek, M, Michel, G. | | Deposit date: | 2014-02-28 | | Release date: | 2015-01-14 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural and Biochemical Characterization of the Laminarina Zglamc[Gh16] from Zobellia Galactanivorans Suggests Preferred Recognition of Branched Laminarin
Acta Crystallogr.,Sect.D, 71, 2015
|
|
1NAZ
 
 | | structure of microgravity-grown oxidized myoglobin mutant YQR (ISS8A) | | Descriptor: | HYDROXIDE ION, Myoglobin, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Miele, A.E, Federici, L, Sciara, G, Draghi, F, Brunori, M, Vallone, B. | | Deposit date: | 2002-11-30 | | Release date: | 2003-06-10 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.04 Å) | | Cite: | Analysis of the effect of microgravity on protein crystal quality: the case of a myoglobin triple mutant.
Acta Crystallogr.,Sect.D, 59, 2003
|
|
8QZH
 
 | | Crystal structure of apo-PptT from Mycobacterium tuberculosis | | Descriptor: | 4'-phosphopantetheinyl transferase PptT, ACETATE ION, MAGNESIUM ION | | Authors: | Gavalda, S, Carivenc, C, Mourey, L, Pedelacq, J.D. | | Deposit date: | 2023-10-27 | | Release date: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Catalytic Cycle of Type II 4'-Phosphopantetheinyl Transferases
Acs Catalysis, 14, 2024
|
|
1NAX
 
 | | Thyroid receptor beta1 in complex with a beta-selective ligand | | Descriptor: | Thyroid hormone receptor beta-1, {3,5-DICHLORO-4-[4-HYDROXY-3-(PROPAN-2-YL)PHENOXY]PHENYL}ACETIC ACID | | Authors: | Ye, L, Li, Y.L, Mellstrom, K, Mellin, C, Bladh, L.G, Koehler, K, Garg, N, Garcia Collazo, A.M, Litten, C, Husman, B, Persson, K, Ljunggren, J, Grover, G, Sleph, P.G, George, R, Malm, J. | | Deposit date: | 2002-11-29 | | Release date: | 2003-06-17 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Thyroid receptor ligands. 1. Agonist ligands selective for the thyroid receptor beta1.
J.Med.Chem., 46, 2003
|
|
1NBL
 
 | |
