6F1S
 
 | | C-terminal domain of CglI restriction endonuclease H subunit | | Descriptor: | 1,2-ETHANEDIOL, CglIIR protein, FORMIC ACID | | Authors: | Tamulaitiene, G, Grigaitis, R, Zaremba, M, Silanskas, A. | | Deposit date: | 2017-11-23 | | Release date: | 2018-02-14 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The H-subunit of the restriction endonuclease CglI contains a prototype DEAD-Z1 helicase-like motor.
Nucleic Acids Res., 46, 2018
|
|
5CRL
 
 | |
6L8W
 
 | | Crystal structure of ugt transferase mutant2 | | Descriptor: | Glycosyltransferase | | Authors: | Li, J, Shan, N, Yang, J.G, Liu, W.D, Sun, Y.X. | | Deposit date: | 2019-11-07 | | Release date: | 2020-04-01 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Efficient O-Glycosylation of Triterpenes Enabled by Protein Engineering of Plant Glycosyltransferase UGT74AC1
Acs Catalysis, 2020
|
|
6L90
 
 | | Crystal structure of ugt transferase enzyme | | Descriptor: | Glycosyltransferase, SULFATE ION | | Authors: | Li, J, Shan, N, Yang, J.G, Liu, W.D, Sun, Y.X. | | Deposit date: | 2019-11-07 | | Release date: | 2020-04-01 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Efficient O-Glycosylation of Triterpenes Enabled by Protein Engineering of Plant Glycosyltransferase UGT74AC1
Acs Catalysis, 2020
|
|
6L8X
 
 | | Crystal structure of Siraitia grosvenorii ugt transferase mutant2 | | Descriptor: | Glycosyltransferase | | Authors: | Li, J, Shan, N, Yang, J.G, Liu, W.D, Sun, Y.X. | | Deposit date: | 2019-11-07 | | Release date: | 2020-04-08 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Efficient O-Glycosylation of Triterpenes Enabled by Protein Engineering of Plant Glycosyltransferase UGT74AC1
Acs Catalysis, 10, 2020
|
|
8ZYM
 
 | | Complex structure of 60 Fab bound to DS2 prefusion F trimer | | Descriptor: | Fusion glycoprotein F0,Expression tag, antibody light chain, antidody heavy chain | | Authors: | Wang, X, Ge, J, Guo, F. | | Deposit date: | 2024-06-18 | | Release date: | 2025-04-30 | | Method: | ELECTRON MICROSCOPY (3.23 Å) | | Cite: | DS2 designer pre-fusion F vaccine induces strong and protective antibody response against RSV infection.
Npj Vaccines, 9, 2024
|
|
6M24
 
 | | Uncommon structural features of rabbit MHC class I (RLA-A1) complexed with rabbit haemorrhagic disease virus (RHDV) derived peptide, VP60-2 | | Descriptor: | Beta-2-microglobulin, RLA class I histocompatibility antigen, alpha chain 19-1, ... | | Authors: | Zhang, Q.X, Liu, K.F, Yue, C, Zhang, D, Lu, D, Xiao, W.L, Liu, P.P, Zhao, Y.Z, Gao, G.L, Ding, C.M, Lyu, J.X, Liu, W.J. | | Deposit date: | 2020-02-26 | | Release date: | 2020-07-08 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Strict Assembly Restriction of Peptides from Rabbit Hemorrhagic Disease Virus Presented by Rabbit Major Histocompatibility Complex Class I Molecule RLA-A1.
J.Virol., 94, 2020
|
|
6M2J
 
 | | Uncommon structural features of rabbit MHC class I (RLA-A1) complexed with rabbit haemorrhagic disease virus (RHDV) derived peptide, VP60-1 | | Descriptor: | Beta-2-microglobulin, RLA class I histocompatibility antigen, alpha chain 19-1, ... | | Authors: | Zhang, Q.X, Liu, K.F, Yue, C, Zhang, D, Lu, D, Xiao, W.L, Liu, P.P, Zhao, Y.Z, Gao, G.L, Ding, C.M, Lyu, J.X, Liu, W.J. | | Deposit date: | 2020-02-27 | | Release date: | 2020-07-08 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Strict Assembly Restriction of Peptides from Rabbit Hemorrhagic Disease Virus Presented by Rabbit Major Histocompatibility Complex Class I Molecule RLA-A1.
J.Virol., 94, 2020
|
|
6M2K
 
 | | Uncommon structural features of rabbit MHC class I (RLA-A1) complexed with rabbit haemorrhagic disease virus (RHDV) derived peptide, VP60-10 | | Descriptor: | Beta-2-microglobulin, RLA class I histocompatibility antigen, alpha chain 19-1, ... | | Authors: | Zhang, Q.X, Liu, K.F, Yue, C, Zhang, D, Lu, D, Xiao, W.L, Liu, P.P, Zhao, Y.Z, Gao, G.L, Ding, C.M, Lyu, J.X, Liu, W.J. | | Deposit date: | 2020-02-27 | | Release date: | 2020-07-08 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Strict Assembly Restriction of Peptides from Rabbit Hemorrhagic Disease Virus Presented by Rabbit Major Histocompatibility Complex Class I Molecule RLA-A1.
J.Virol., 94, 2020
|
|
7F7E
 
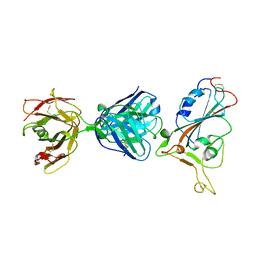 | | SARS-CoV-2 S protein RBD in complex with A5-10 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of A5-10 Fab, Light chain of A5-10 Fab, ... | | Authors: | Dou, Y, Wang, X, Wang, K, Liu, P, Lu, B. | | Deposit date: | 2021-06-29 | | Release date: | 2022-02-02 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Etesevimab in combination with JS026 neutralizing SARS-CoV-2 and its variants.
Emerg Microbes Infect, 11, 2022
|
|
8IAK
 
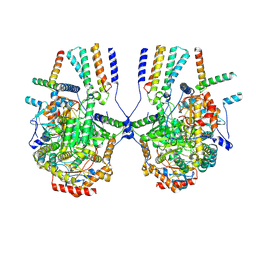 | | Cryo-EM structure of the yeast SPT-ORM2 (ORM2-S3A-N71A) complex | | Descriptor: | Long chain base biosynthesis protein 1,Serine palmitoyltransferase 1, Protein ORM2, Serine palmitoyltransferase 2, ... | | Authors: | Xie, T, Gong, X. | | Deposit date: | 2023-02-08 | | Release date: | 2024-02-14 | | Last modified: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Collaborative regulation of yeast SPT-Orm2 complex by phosphorylation and ceramide.
Cell Rep, 43, 2024
|
|
8IAJ
 
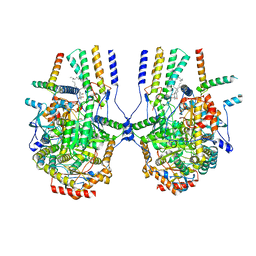 | | Cryo-EM structure of the yeast SPT-ORM2 (ORM2-S3A) complex | | Descriptor: | N-[(2S,3R,4E)-1,3-dihydroxyoctadec-4-en-2-yl]tetracosanamide, PYRIDOXAL-5'-PHOSPHATE, Protein ORM2, ... | | Authors: | Xie, T, Gong, X. | | Deposit date: | 2023-02-08 | | Release date: | 2024-02-14 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Collaborative regulation of yeast SPT-Orm2 complex by phosphorylation and ceramide.
Cell Rep, 43, 2024
|
|
8IAM
 
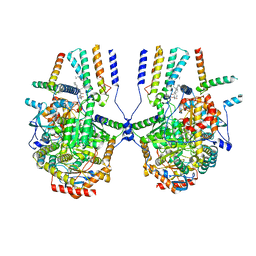 | | Cryo-EM structure of the yeast SPT-ORM2 (ORM2-S3D) complex | | Descriptor: | Chimera of Long chain base biosynthesis protein 1 and Serine palmitoyltransferase 1, N-[(2S,3R,4E)-1,3-dihydroxyoctadec-4-en-2-yl]tetracosanamide, PYRIDOXAL-5'-PHOSPHATE, ... | | Authors: | Xie, T, Gong, X. | | Deposit date: | 2023-02-08 | | Release date: | 2024-02-14 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Collaborative regulation of yeast SPT-Orm2 complex by phosphorylation and ceramide.
Cell Rep, 43, 2024
|
|
8QSV
 
 | | Crystal structure of SPOUT1/CENP-32 bound to SAM | | Descriptor: | Putative methyltransferase C9orf114, S-ADENOSYLMETHIONINE | | Authors: | Jeyaprakash, A.A, Abad, M.A. | | Deposit date: | 2023-10-11 | | Release date: | 2024-10-23 | | Last modified: | 2025-05-28 | | Method: | X-RAY DIFFRACTION (2.62 Å) | | Cite: | RNA methyltransferase SPOUT1/CENP-32 links mitotic spindle organization with the neurodevelopmental disorder SpADMiSS.
Nat Commun, 16, 2025
|
|
8QSU
 
 | | Crystal structure of SPOUT1/CENP-32 bound to SAH | | Descriptor: | Putative methyltransferase C9orf114, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Jeyaprakash, A.A, Abad, M.A. | | Deposit date: | 2023-10-11 | | Release date: | 2024-10-23 | | Last modified: | 2025-05-28 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | RNA methyltransferase SPOUT1/CENP-32 links mitotic spindle organization with the neurodevelopmental disorder SpADMiSS.
Nat Commun, 16, 2025
|
|
8QSW
 
 | |
7ANZ
 
 | | Structure of the Candida albicans gamma-Tubulin Small Complex | | Descriptor: | Spindle pole body component, Tubulin gamma chain | | Authors: | Zupa, E, Pfeffer, S. | | Deposit date: | 2020-10-13 | | Release date: | 2020-11-04 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | The cryo-EM structure of a gamma-TuSC elucidates architecture and regulation of minimal microtubule nucleation systems.
Nat Commun, 11, 2020
|
|
3DEZ
 
 | | Crystal structure of Orotate phosphoribosyltransferase from Streptococcus mutans | | Descriptor: | Orotate phosphoribosyltransferase, SULFATE ION | | Authors: | Liu, C.P, Gao, Z.Q, Hou, H.F, Li, L.F, Su, X.D, Dong, Y.H. | | Deposit date: | 2008-06-11 | | Release date: | 2009-06-16 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of orotate phosphoribosyltransferase from the caries pathogen Streptococcus mutans
Acta Crystallogr.,Sect.F, 66, 2010
|
|
6M2Z
 
 | | Crystal structure of a formolase, BFD variant M3 from Pseudomonas putida | | Descriptor: | Benzoylformate decarboxylase, MAGNESIUM ION, THIAMINE DIPHOSPHATE | | Authors: | Wei, H.L, Liu, W.D, Li, T.Z, Zhu, L.L. | | Deposit date: | 2020-03-02 | | Release date: | 2021-03-03 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Totally atom-economical synthesis of lactic acid from formaldehyde: combined bio-carboligation and chemo-rearrangement without the isolation of intermediate.
Green Chem, 22, 2020
|
|
6M2Y
 
 | | Crystal structure of a formolase, BFD variant M6 from Pseudomonas putida | | Descriptor: | Benzoylformate decarboxylase, MAGNESIUM ION, THIAMINE DIPHOSPHATE | | Authors: | Wei, H.L, Liu, W.D, Li, T.Z, Zhu, L.L. | | Deposit date: | 2020-03-02 | | Release date: | 2021-03-03 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Totally atom-economical synthesis of lactic acid from formaldehyde: combined bio-carboligation and chemo-rearrangement without the isolation of intermediate.
Green Chem, 22, 2020
|
|
7F7H
 
 | | SARS-CoV-2 S protein RBD in complex with A8-1 Fab | | Descriptor: | Heavy chain of A8-1 Fab, Light chain of A8-1 Fab, Spike glycoprotein S1 | | Authors: | Dou, Y, Wang, X, Wang, K, Liu, P, Lu, B. | | Deposit date: | 2021-06-29 | | Release date: | 2022-06-15 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (3.19 Å) | | Cite: | High throughput isolation of potent neutralizing antibodies from convalescent COVID-19 patients.
To Be Published
|
|
8E80
 
 | | Structure of LRRK2-CHK1 10-pt. mutant complex with heteroaryl-1H-indazole LRRK2 inhibitor 14 | | Descriptor: | 2-[(1r,4r)-2-{(6P)-6-[(6M)-6-(1H-pyrazol-5-yl)-1H-indazol-1-yl]pyrimidin-4-yl}-2-azabicyclo[2.1.1]hexan-4-yl]propan-2-ol, Serine/threonine-protein kinase Chk1 | | Authors: | Palte, R.L. | | Deposit date: | 2022-08-25 | | Release date: | 2023-02-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Discovery and Optimization of Potent, Selective, and Brain-Penetrant 1-Heteroaryl-1 H -Indazole LRRK2 Kinase Inhibitors for the Treatment of Parkinson's Disease.
J.Med.Chem., 65, 2022
|
|
8E81
 
 | | Structure of LRRK2-CHK1 10-pt. mutant complex with heteroaryl-1H-indazole LRRK2 inhibitor 25 | | Descriptor: | (1S)-1-[(1P)-1-{6-[(3R)-3-(2-hydroxypropan-2-yl)pyrrolidin-1-yl]pyrimidin-4-yl}-1H-indazol-6-yl]spiro[2.2]pentane-1-carbonitrile, Serine/threonine-protein kinase Chk1 | | Authors: | Palte, R.L. | | Deposit date: | 2022-08-25 | | Release date: | 2023-02-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Discovery and Optimization of Potent, Selective, and Brain-Penetrant 1-Heteroaryl-1 H -Indazole LRRK2 Kinase Inhibitors for the Treatment of Parkinson's Disease.
J.Med.Chem., 65, 2022
|
|
9C6O
 
 | |
9D32
 
 | |
