3I3C
 
 | | Crystal Structural of CBX5 Chromo Shadow Domain | | Descriptor: | Chromobox protein homolog 5, SODIUM ION | | Authors: | Amaya, M.F, Li, Z, Li, Y, Kozieradzki, I, Edwards, A.M, Arrowsmith, C.H, Weigelt, J, Bountra, C, Bochkarev, A, Min, J, Ouyang, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-06-30 | | Release date: | 2009-08-18 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Crystal Structural of CBX5 Chromo Shadow Domain
To be Published
|
|
6VSI
 
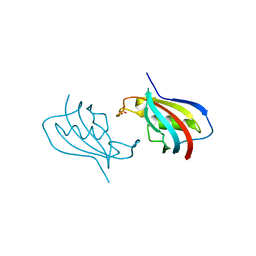 | | Crystal structure of FKBP12 of Candida auris | | Descriptor: | Peptidylprolyl isomerase, SULFATE ION | | Authors: | Li, Z, Li, H, Hernandez, G, LeMaster, D. | | Deposit date: | 2020-02-11 | | Release date: | 2020-04-01 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Crystal structure and transient dimerization for the FKBP12 protein from the pathogenic fungus Candida auris.
Biochem.Biophys.Res.Commun., 525, 2020
|
|
5BKH
 
 | |
5TJX
 
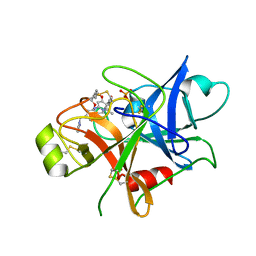 | | Structure of human plasma kallikrein | | Descriptor: | (8E)-3-amino-1-methyl-15-[(1H-pyrazol-1-yl)methyl]-7,10,11,12,24,25-hexahydro-6H,18H,23H-19,22-(metheno)pyrido[4,3-j][1,9,13,17,18]benzodioxatriazacyclohenicosin-23-one, PHOSPHATE ION, Plasma kallikrein | | Authors: | Partridge, J.R, Choy, R.M, Li, Z. | | Deposit date: | 2016-10-05 | | Release date: | 2016-12-21 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.408 Å) | | Cite: | Structure-Guided Design of Novel, Potent, and Selective Macrocyclic Plasma Kallikrein Inhibitors.
ACS Med Chem Lett, 8, 2017
|
|
6MX6
 
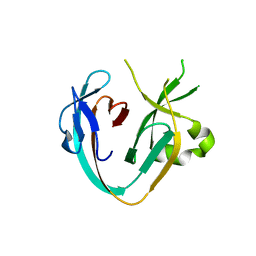 | | The Prp8 intein of Cryptococcus neoformans | | Descriptor: | Pre-mRNA-processing-splicing factor 8 | | Authors: | Li, Z, Li, H. | | Deposit date: | 2018-10-30 | | Release date: | 2019-10-23 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.749 Å) | | Cite: | Spliceosomal Prp8 intein at the crossroads of protein and RNA splicing.
Plos Biol., 17, 2019
|
|
3QII
 
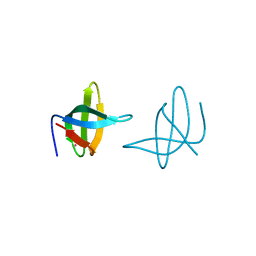 | | Crystal structure of tudor domain 2 of human PHD finger protein 20 | | Descriptor: | PHD finger protein 20, UNKNOWN ATOM OR ION | | Authors: | Li, Z, Tempel, W, Wernimont, A.K, Chao, X, Bian, C, Lam, R, Crombet, L, Bountra, C, Weigelt, J, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2011-01-27 | | Release date: | 2011-02-09 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structures of the Tudor domains of human PHF20 reveal novel structural variations on the Royal Family of proteins.
Febs Lett., 586, 2012
|
|
6XMG
 
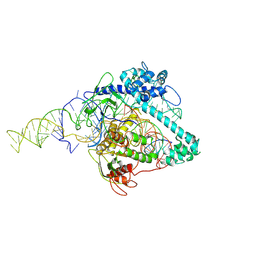 | | Cryo-EM structure of Cas12g ternary complex | | Descriptor: | CRISPR-Cas, RNA (130-MER), RNA (5'-R(P*UP*UP*AP*AP*UP*GP*CP*GP*GP*UP*AP*GP*UP*UP*UP*AP*UP*CP*AP*CP*AP*GP*UP*U)-3'), ... | | Authors: | Chang, L, Li, Z, Zhang, H. | | Deposit date: | 2020-06-30 | | Release date: | 2021-01-13 | | Last modified: | 2025-05-14 | | Method: | ELECTRON MICROSCOPY (4.8 Å) | | Cite: | Cryo-EM structure of the RNA-guided ribonuclease Cas12g.
Nat.Chem.Biol., 17, 2021
|
|
5I0A
 
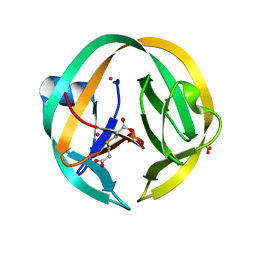 | | RecA mini intein in complex with cisplatin | | Descriptor: | 3,3',3''-phosphanetriyltripropanoic acid, Cisplatin, Intein, ... | | Authors: | Li, Z, Zhang, J, Li, H.M. | | Deposit date: | 2016-02-03 | | Release date: | 2016-09-21 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural insights into protein splicing inhibition by platinum therapeutics as potential anti-microbials
To be published
|
|
6XMF
 
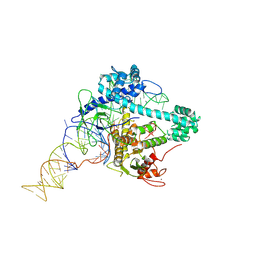 | |
6X8Z
 
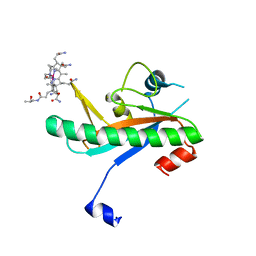 | | Crystal structure of N-truncated human B12 chaperone CblD(C262S)-thiolato-cob(III)alamin complex (108-296) | | Descriptor: | COBALAMIN, Methylmalonic aciduria and homocystinuria type D protein, mitochondrial | | Authors: | Mascarenhas, R, Li, Z, Koutmos, M, Banerjee, R. | | Deposit date: | 2020-06-02 | | Release date: | 2020-09-16 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | An Interprotein Co-S Coordination Complex in the B 12 -Trafficking Pathway.
J.Am.Chem.Soc., 142, 2020
|
|
1GGO
 
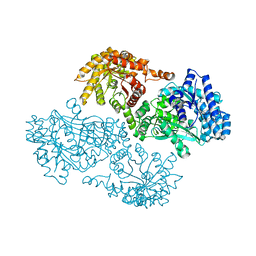 | | T453A MUTANT OF PYRUVATE, PHOSPHATE DIKINASE | | Descriptor: | PROTEIN (PYRUVATE, PHOSPHATE DIKINASE), SULFATE ION | | Authors: | Li, Z, Herzberg, O. | | Deposit date: | 2000-08-29 | | Release date: | 2001-01-10 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Identification of domain-domain docking sites within Clostridium symbiosum pyruvate phosphate dikinase by amino acid replacement.
J.Biol.Chem., 275, 2000
|
|
5XJ7
 
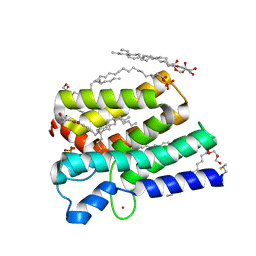 | | Crystal structure of PlsY (YgiH), an integral membrane glycerol 3-phosphate acyltransferase - the acyl phosphate form | | Descriptor: | (2S)-2,3-DIHYDROXYPROPYL(7Z)-PENTADEC-7-ENOATE, Glycerol-3-phosphate acyltransferase, PHOSPHATE ION, ... | | Authors: | Tang, Y, Li, Z, Li, D. | | Deposit date: | 2017-04-30 | | Release date: | 2017-12-06 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.772 Å) | | Cite: | Structural insights into the committed step of bacterial phospholipid biosynthesis.
Nat Commun, 8, 2017
|
|
8EWG
 
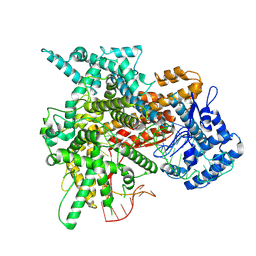 | | Cryo-EM structure of a riboendonclease | | Descriptor: | CRISPR-associated endonuclease Cas9, RNA (56-MER) | | Authors: | Li, Z, Wang, F. | | Deposit date: | 2022-10-23 | | Release date: | 2023-08-30 | | Last modified: | 2025-05-21 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural Basis for the Ribonuclease Activity of a Thermostable CRISPR-Cas13a from Thermoclostridium caenicola.
J.Mol.Biol., 435, 2023
|
|
7KMK
 
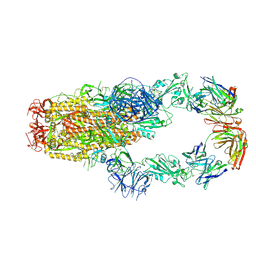 | | cryo-EM structure of SARS-CoV-2 spike in complex with Fab 15033-7, two RBDs bound | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Fab 15033-7 heavy chain, ... | | Authors: | Li, Z, Rini, J.M. | | Deposit date: | 2020-11-03 | | Release date: | 2021-02-10 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Tetravalent SARS-CoV-2 Neutralizing Antibodies Show Enhanced Potency and Resistance to Escape Mutations.
J.Mol.Biol., 433, 2021
|
|
7KLG
 
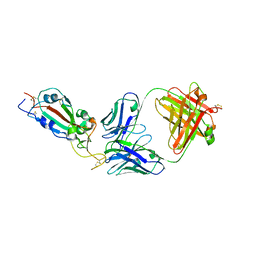 | | SARS-CoV-2 RBD in complex with Fab 15033 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Fab 15033 heavy chain, Fab 15033 light chain, ... | | Authors: | Li, Z, Rini, J.M. | | Deposit date: | 2020-10-30 | | Release date: | 2021-02-10 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Tetravalent SARS-CoV-2 Neutralizing Antibodies Show Enhanced Potency and Resistance to Escape Mutations.
J.Mol.Biol., 433, 2021
|
|
7KML
 
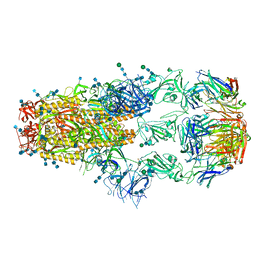 | | cryo-EM structure of SARS-CoV-2 spike in complex with Fab 15033-7, three RBDs bound | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Fab 15033-7 heavy chain, ... | | Authors: | Li, Z, Rini, J.M. | | Deposit date: | 2020-11-03 | | Release date: | 2021-02-10 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Tetravalent SARS-CoV-2 Neutralizing Antibodies Show Enhanced Potency and Resistance to Escape Mutations.
J.Mol.Biol., 433, 2021
|
|
7KLH
 
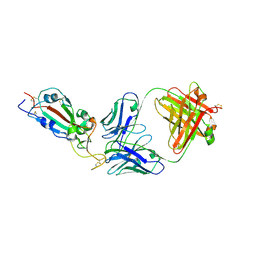 | | SARS-CoV-2 RBD in complex with Fab 15033-7 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Fab 15033-7 heavy chain, Fab 15033-7 light chain, ... | | Authors: | Li, Z, Rini, J.M. | | Deposit date: | 2020-10-30 | | Release date: | 2021-02-10 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Tetravalent SARS-CoV-2 Neutralizing Antibodies Show Enhanced Potency and Resistance to Escape Mutations.
J.Mol.Biol., 433, 2021
|
|
2MAY
 
 | |
7X77
 
 | | Ectodomain structure of per os infectivity factor 5 | | Descriptor: | Per os infectivity factor 5 | | Authors: | Cao, S, Li, Z, Fu, Y. | | Deposit date: | 2022-03-09 | | Release date: | 2022-06-22 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural Characterization of Per Os Infectivity Factor 5 (PIF5) Reveals the Essential Role of Intramolecular Interactions in Baculoviral Oral Infectivity.
J.Virol., 96, 2022
|
|
6U7E
 
 | | HCoV-229E RBD Class III in complex with human APN | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Aminopeptidase N, ... | | Authors: | Tomlinson, A.C.A, Li, Z, Rini, J.M. | | Deposit date: | 2019-09-02 | | Release date: | 2019-11-13 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The human coronavirus HCoV-229E S-protein structure and receptor binding.
Elife, 8, 2019
|
|
8E2L
 
 | | Structure of Lates calcarifer Twinkle helicase with ATP and DNA | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, DNA (5'-D(P*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*T)-3'), MAGNESIUM ION, ... | | Authors: | Gao, Y, Li, Z. | | Deposit date: | 2022-08-15 | | Release date: | 2022-11-02 | | Last modified: | 2025-05-28 | | Method: | ELECTRON MICROSCOPY (3.51 Å) | | Cite: | Structural and dynamic basis of DNA capture and translocation by mitochondrial Twinkle helicase.
Nucleic Acids Res., 50, 2022
|
|
6U7G
 
 | | HCoV-229E RBD Class V in complex with human APN | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Aminopeptidase N, ... | | Authors: | Tomlinson, A, Li, Z, Rini, J.M. | | Deposit date: | 2019-09-02 | | Release date: | 2019-11-13 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | The human coronavirus HCoV-229E S-protein structure and receptor binding.
Elife, 8, 2019
|
|
6U7F
 
 | | HCoV-229E RBD Class IV in complex with human APN | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Aminopeptidase N, ... | | Authors: | Tomlinson, A.C.A, Li, Z, Rini, J.M. | | Deposit date: | 2019-09-02 | | Release date: | 2019-11-13 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | The human coronavirus HCoV-229E S-protein structure and receptor binding.
Elife, 8, 2019
|
|
6UVN
 
 | | CryoEM structure of VcCascasde-TniQ complex | | Descriptor: | Cas6, Cas7, Cas8/5, ... | | Authors: | Chang, L, Li, Z, Zhang, H. | | Deposit date: | 2019-11-03 | | Release date: | 2020-01-29 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Cryo-EM structure of a type I-F CRISPR RNA guided surveillance complex bound to transposition protein TniQ.
Cell Res., 30, 2020
|
|
8CUS
 
 | |
