2Y5B
 
 | | Structure of USP21 in complex with linear diubiquitin-aldehyde | | Descriptor: | SULFATE ION, UBIQUITIN, UBIQUITIN CARBOXYL-TERMINAL HYDROLASE 21, ... | | Authors: | Ye, Y, Akutsu, M, Reyes-Turcu, F, Enchev, R.I, Wilkinson, K.D, Komander, D. | | Deposit date: | 2011-01-12 | | Release date: | 2012-01-25 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Polyubiquitin Binding and Cross-Reactivity in the Usp Domain Deubiquitinase Usp21.
Embo Rep., 12, 2011
|
|
5AF6
 
 | | Structure of Lys33-linked diUb bound to Trabid NZF1 | | Descriptor: | TRABID, UBIQUITIN, ZINC ION | | Authors: | Michel, M.A, Elliott, P.R, Swatek, K.N, Simicek, M, Pruneda, J.N, Wagstaff, J.L, Freund, S.M.V, Komander, D. | | Deposit date: | 2015-01-19 | | Release date: | 2015-03-25 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Assembly and Specific Recognition of K29- and K33-Linked Polyubiquitin.
Mol.Cell, 58, 2015
|
|
5AF5
 
 | | Structure of Lys33-linked triUb S.G. P 212121 | | Descriptor: | POLYUBIQUITIN-C | | Authors: | Michel, M.A, Elliott, P.R, Swatek, K.N, Simicek, M, Pruneda, J.N, Wagstaff, J.L, Freund, S.M.V, Komander, D. | | Deposit date: | 2015-01-19 | | Release date: | 2015-03-25 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Assembly and Specific Recognition of K29- and K33-Linked Polyubiquitin.
Mol.Cell, 58, 2015
|
|
5AF4
 
 | | Structure of Lys33-linked diUb | | Descriptor: | GLYCEROL, SULFATE ION, UBIQUITIN | | Authors: | Michel, M.A, Elliott, P.R, Swatek, K.N, Simicek, M, Pruneda, J.N, Wagstaff, J.L, Freund, S.M.V, Komander, D. | | Deposit date: | 2015-01-19 | | Release date: | 2015-03-25 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Assembly and Specific Recognition of K29- and K33-Linked Polyubiquitin.
Mol.Cell, 58, 2015
|
|
5CAW
 
 | |
5OE7
 
 | |
1UPQ
 
 | | Crystal structure of the pleckstrin homology (PH) domain of PEPP1 | | Descriptor: | PEPP1, SULFATE ION | | Authors: | Milburn, C.C, Komander, D, Deak, M, Alessi, D.R, van Aalten, D.M.F. | | Deposit date: | 2003-10-09 | | Release date: | 2004-10-07 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Crystal Structure of the Pleckstrin Homology Domain of Pepp1
To be Published
|
|
1UPR
 
 | | Crystal structure of the PEPP1 pleckstrin homology domain in complex with Inositol 1,3,4,5-tetrakisphosphate | | Descriptor: | INOSITOL-(1,3,4,5)-TETRAKISPHOSPHATE, PLECKSTRIN HOMOLOGY DOMAIN-CONTAINING FAMILY A MEMBER 4 | | Authors: | Milburn, C.C, Komander, D, Deak, M, Alessi, D.R, Van Aalten, D.M.F. | | Deposit date: | 2003-10-10 | | Release date: | 2004-10-20 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Crystal Structure of the Pleckstrin Homology Domain of Pepp1
To be Published
|
|
3ZNV
 
 | | Crystal structure of the OTU domain of OTULIN at 1.3 Angstroms. | | Descriptor: | CALCIUM ION, CHLORIDE ION, GLYCEROL, ... | | Authors: | Keusekotten, K, Elliott, P.R, Glockner, L, Kulathu, Y, Wauer, T, Krappmann, D, Hofmann, K, Komander, D. | | Deposit date: | 2013-02-18 | | Release date: | 2013-06-26 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Otulin Antagonizes Lubac Signaling by Specifically Hydrolyzing met1-Linked Polyubiquitin.
Cell(Cambridge,Mass.), 153, 2013
|
|
3ZNZ
 
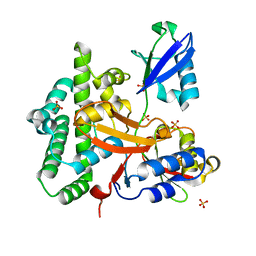 | | Crystal structure of OTULIN OTU domain (C129A) in complex with Met1- di ubiquitin | | Descriptor: | POLYUBIQUITIN-C, PROTEIN FAM105B, SULFATE ION | | Authors: | Keusekotten, K, Elliott, P.R, Glockner, L, Kulathu, Y, Wauer, T, Krappmann, D, Hofmann, K, Komander, D. | | Deposit date: | 2013-02-18 | | Release date: | 2013-06-26 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Otulin Antagonizes Lubac Signaling by Specifically Hydrolyzing met1-Linked Polyubiquitin.
Cell(Cambridge,Mass.), 153, 2013
|
|
3ZJD
 
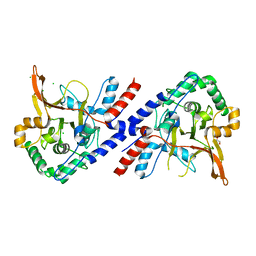 | | A20 OTU domain in reduced, active state at 1.87 A resolution | | Descriptor: | 1,2-ETHANEDIOL, A20P50, CHLORIDE ION | | Authors: | Kulathu, Y, Garcia, F.J, Mevissen, T.E.T, Busch, M, Arnaudo, N, Carroll, K.S, Barford, D, Komander, D. | | Deposit date: | 2013-01-17 | | Release date: | 2013-03-06 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Regulation of A20 and Other Otu Deubiquitinases by Reversible Oxidation
Nat.Commun., 4, 2013
|
|
3ZJF
 
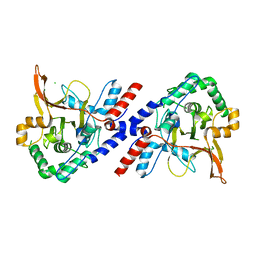 | | A20 OTU domain with irreversibly oxidised Cys103 from 270 min H2O2 soak. | | Descriptor: | A20P50, CHLORIDE ION | | Authors: | Kulathu, Y, Garcia, F.J, Mevissen, T.E.T, Busch, M, Arnaudo, N, Carroll, K.S, Barford, D, Komander, D. | | Deposit date: | 2013-01-17 | | Release date: | 2013-03-06 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Regulation of A20 and Other Otu Deubiquitinases by Reversible Oxidation
Nat.Commun., 4, 2013
|
|
3ZNX
 
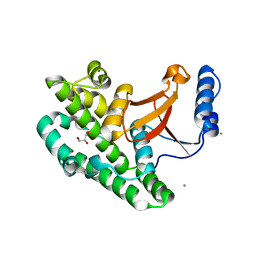 | | Crystal structure of the OTU domain of OTULIN D336A mutant | | Descriptor: | CALCIUM ION, CHLORIDE ION, GLYCEROL, ... | | Authors: | Keusekotten, K, Elliott, P.R, Glockner, L, Kulathu, Y, Wauer, T, Krappmann, D, Hofmann, K, Komander, D. | | Deposit date: | 2013-02-18 | | Release date: | 2013-06-26 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Otulin Antagonizes Lubac Signaling by Specifically Hydrolyzing met1-Linked Polyubiquitin.
Cell(Cambridge,Mass.), 153, 2013
|
|
3ZJG
 
 | | A20 OTU domain with irreversibly oxidised Cys103 from 60 min H2O2 soak. | | Descriptor: | CHLORIDE ION, TUMOR NECROSIS FACTOR ALPHA-INDUCED PROTEIN 3 | | Authors: | Kulathu, Y, Garcia, F.J, Mevissen, T.E.T, Busch, M, Arnaudo, N, Carroll, K.S, Barford, D, Komander, D. | | Deposit date: | 2013-01-17 | | Release date: | 2013-03-06 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Regulation of A20 and Other Otu Deubiquitinases by Reversible Oxidation
Nat.Commun., 4, 2013
|
|
3ZJE
 
 | | A20 OTU domain in reversibly oxidised (SOH) state | | Descriptor: | 1,2-ETHANEDIOL, A20P50, CHLORIDE ION | | Authors: | Kulathu, Y, Garcia, F.J, Mevissen, T.E.T, Busch, M, Arnaudo, N, Carroll, K.S, Barford, D, Komander, D. | | Deposit date: | 2013-01-17 | | Release date: | 2013-03-06 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Regulation of A20 and Other Otu Deubiquitinases by Reversible Oxidation
Nat.Commun., 4, 2013
|
|
5OHV
 
 | | K33-specific affimer bound to K33 diUb | | Descriptor: | GLYCEROL, K33-specific affimer, SULFATE ION, ... | | Authors: | Michel, M.A, Komander, D. | | Deposit date: | 2017-07-18 | | Release date: | 2017-10-04 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.801 Å) | | Cite: | Ubiquitin Linkage-Specific Affimers Reveal Insights into K6-Linked Ubiquitin Signaling.
Mol. Cell, 68, 2017
|
|
5OHM
 
 | | K33-specific affimer bound to K33 diUb | | Descriptor: | K33-specific affimer, POLYETHYLENE GLYCOL (N=34), Polyubiquitin-C | | Authors: | Michel, M.A, Komander, D. | | Deposit date: | 2017-07-17 | | Release date: | 2017-10-04 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | Ubiquitin Linkage-Specific Affimers Reveal Insights into K6-Linked Ubiquitin Signaling.
Mol. Cell, 68, 2017
|
|
5OHN
 
 | |
5OHL
 
 | | K6-specific affimer bound to K6 diUb | | Descriptor: | GLYCEROL, K6-specific affimer, POLYETHYLENE GLYCOL (N=34), ... | | Authors: | Michel, M.A, Komander, D. | | Deposit date: | 2017-07-17 | | Release date: | 2017-10-04 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Ubiquitin Linkage-Specific Affimers Reveal Insights into K6-Linked Ubiquitin Signaling.
Mol. Cell, 68, 2017
|
|
5OXI
 
 | | C-terminally retracted ubiquitin L67S mutant | | Descriptor: | SULFATE ION, Ubiquitin L67S mutant | | Authors: | Gladkova, C.G, Schubert, A.F, Wagstaff, J.L, Pruneda, J.N, Freund, S.M.V, Komander, D. | | Deposit date: | 2017-09-06 | | Release date: | 2017-11-22 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | An invisible ubiquitin conformation is required for efficient phosphorylation by PINK1.
EMBO J., 36, 2017
|
|
5OXH
 
 | | C-terminally retracted ubiquitin T66V/L67N mutant | | Descriptor: | SULFATE ION, Ubiquitin T66V/L67N mutant | | Authors: | Gladkova, C, Schubert, A.F, Wagstaff, J.L, Pruneda, J.P, Freund, S.M.V, Komander, D. | | Deposit date: | 2017-09-06 | | Release date: | 2017-11-22 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.601 Å) | | Cite: | An invisible ubiquitin conformation is required for efficient phosphorylation by PINK1.
EMBO J., 36, 2017
|
|
5OHP
 
 | |
5OHK
 
 | | Crystal structure of USP30 in covalent complex with ubiquitin propargylamide (high resolution) | | Descriptor: | Polyubiquitin-B, Ubiquitin carboxyl-terminal hydrolase 30,Ubiquitin carboxyl-terminal hydrolase 30,Ubiquitin carboxyl-terminal hydrolase 30, ZINC ION, ... | | Authors: | Gersch, M, Komander, D. | | Deposit date: | 2017-07-17 | | Release date: | 2017-09-20 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Mechanism and regulation of the Lys6-selective deubiquitinase USP30.
Nat. Struct. Mol. Biol., 24, 2017
|
|
5NGF
 
 | | Crystal structure of USP7 in complex with the covalent inhibitor, FT827 | | Descriptor: | 1,2-ETHANEDIOL, Ubiquitin carboxyl-terminal hydrolase 7, ~{N}-[2-[4-[4-[(1-methyl-4-oxidanylidene-pyrazolo[3,4-d]pyrimidin-5-yl)methyl]-4-oxidanyl-piperidin-1-yl]carbonylphenyl]phenyl]ethanesulfonamide | | Authors: | Krajewski, W.W, Turnbull, A.P, Ioannidis, S, Kessler, B.M, Komander, D. | | Deposit date: | 2017-03-17 | | Release date: | 2017-10-18 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Molecular basis of USP7 inhibition by selective small-molecule inhibitors.
Nature, 550, 2017
|
|
5NGE
 
 | | Crystal structure of USP7 in complex with the non-covalent inhibitor, FT671 | | Descriptor: | 5-[[1-[(3~{S})-4,4-bis(fluoranyl)-3-(3-fluoranylpyrazol-1-yl)butanoyl]-4-oxidanyl-piperidin-4-yl]methyl]-1-(4-fluorophenyl)pyrazolo[3,4-d]pyrimidin-4-one, Ubiquitin carboxyl-terminal hydrolase 7 | | Authors: | Turnbull, A.P, Krajewski, W.W, Ioannidis, S, Kessler, B.M, Komander, D. | | Deposit date: | 2017-03-17 | | Release date: | 2017-10-18 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Molecular basis of USP7 inhibition by selective small-molecule inhibitors.
Nature, 550, 2017
|
|
