6N7X
 
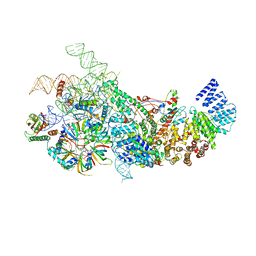 | | S. cerevisiae U1 snRNP | | Descriptor: | 56 kDa U1 small nuclear ribonucleoprotein component, Pre-mRNA-processing factor 39, Protein NAM8, ... | | Authors: | Li, X, Liu, S, Jiang, J, Zhang, L, Espinosa, S, Hill, R.C, Hansen, K.C, Zhou, Z.H, Zhao, R. | | Deposit date: | 2018-11-28 | | Release date: | 2019-07-24 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | CryoEM structure of Saccharomyces cerevisiae U1 snRNP offers insight into alternative splicing.
Nat Commun, 8, 2017
|
|
6OSL
 
 | |
1IIM
 
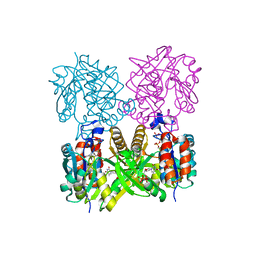 | | thymidylyltransferase complexed with TTP | | Descriptor: | THYMIDINE-5'-TRIPHOSPHATE, glucose-1-phosphate thymidylyltransferase | | Authors: | Barton, W.A, Lesniak, J, Biggins, J.B, Jeffrey, P.D, Jiang, J, Rajashankar, K.R, Thorson, J.S, Nikolov, D.B. | | Deposit date: | 2001-04-23 | | Release date: | 2001-05-09 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure, mechanism and engineering of a nucleotidylyltransferase as a first step toward glycorandomization.
Nat.Struct.Biol., 8, 2001
|
|
8TEL
 
 | |
8TEH
 
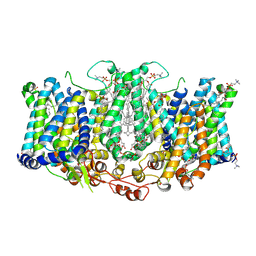 | | Cryo-EM structure of Arabidopsis thaliana Bor1 in lipid nanodiscs | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, Boron transporter 1 | | Authors: | Jiang, Y, Jiang, J. | | Deposit date: | 2023-07-06 | | Release date: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (2.3 Å) | | Cite: | Structure of borate transporter Bor1 reveals a novel auto-inhibition mechanism for
the SLC4 family
To Be Published
|
|
8TEG
 
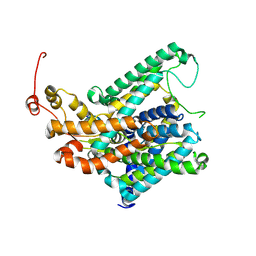 | |
8TEJ
 
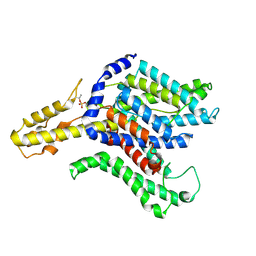 | |
8TEM
 
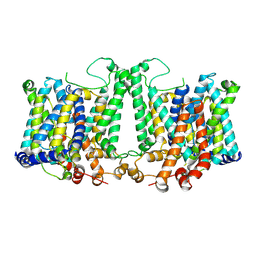 | |
8TEI
 
 | |
8TEN
 
 | |
1IIN
 
 | | thymidylyltransferase complexed with UDP-glucose | | Descriptor: | URIDINE-5'-DIPHOSPHATE-GLUCOSE, glucose-1-phosphate thymidylyltransferase | | Authors: | Barton, W.A, Lesniak, J, Biggins, J.B, Jeffrey, P.D, Jiang, J, Rajashankar, K.R, Thorson, J.S, Nikolov, D.B. | | Deposit date: | 2001-04-23 | | Release date: | 2001-05-09 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure, mechanism and engineering of a nucleotidylyltransferase as a first step toward glycorandomization.
Nat.Struct.Biol., 8, 2001
|
|
7LC9
 
 | |
6TYI
 
 | | ExbB-ExbD complex in MSP1E3D1 nanodisc | | Descriptor: | (1S)-2-{[(2-AMINOETHOXY)(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL STEARATE, (1S)-2-{[{[(2R)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL STEARATE, Biopolymer transport protein ExbB, ... | | Authors: | Celia, H, Botos, I, Jiang, J, Buchanan, S.K. | | Deposit date: | 2019-08-09 | | Release date: | 2019-10-16 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Cryo-EM structure of the bacterial Ton motor subcomplex ExbB-ExbD provides information on structure and stoichiometry.
Commun Biol, 2, 2019
|
|
5TKE
 
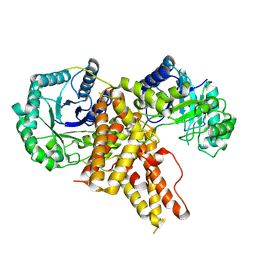 | | Crystal Structure of Eukaryotic Hydrolase | | Descriptor: | O-GlcNAcase: chimera construct | | Authors: | Li, B, Jiang, J. | | Deposit date: | 2016-10-06 | | Release date: | 2017-03-15 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.481 Å) | | Cite: | Structures of human O-GlcNAcase and its complexes reveal a new substrate recognition mode.
Nat. Struct. Mol. Biol., 24, 2017
|
|
8SDU
 
 | | Structure of rat organic anion transporter 1 (OAT1) | | Descriptor: | Solute carrier family 22 member 6 | | Authors: | Dou, T, Jiang, J. | | Deposit date: | 2023-04-07 | | Release date: | 2023-10-18 | | Last modified: | 2023-11-22 | | Method: | ELECTRON MICROSCOPY (2.05 Å) | | Cite: | The substrate and inhibitor binding mechanism of polyspecific transporter OAT1 revealed by high-resolution cryo-EM.
Nat.Struct.Mol.Biol., 30, 2023
|
|
8SDZ
 
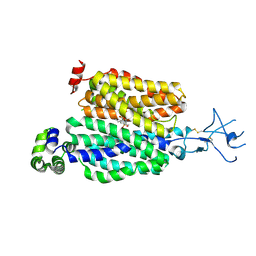 | |
8SDY
 
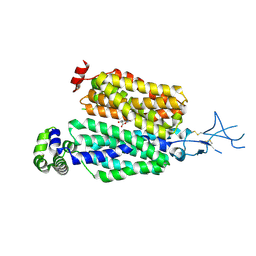 | |
6CI2
 
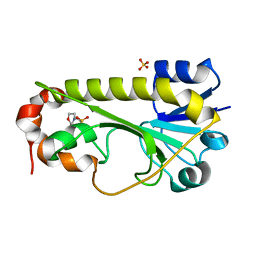 | | Crystal structure of the formyltransferase PseJ from Anoxybacillus kamchatkensis | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, SULFATE ION, formyltransferase PseJ | | Authors: | Reimer, J.M, Jiang, J, Harb, I, Schmeing, T.M. | | Deposit date: | 2018-02-23 | | Release date: | 2018-10-17 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Structural Insight into a Novel Formyltransferase and Evolution to a Nonribosomal Peptide Synthetase Tailoring Domain.
ACS Chem. Biol., 13, 2018
|
|
6E37
 
 | | O-GlcNAc Transferase in complex with covalent inhibitor | | Descriptor: | (2S,3R,4R,5S,6R)-3-[(2E)-but-2-enoylamino]-4,5-dihydroxy-6-(hydroxymethyl)tetrahydro-2H-thiopyran-2-yl [(2R,3S,4R,5R)-5-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)-3,4-dihydroxytetrahydrofuran-2-yl]methyl dihydrogen diphosphate (non-preferred name), O-GlcNAc transferase subunit p110, TYR-PRO-GLY-GLY-SER-THR-PRO-VAL-SER-SER-ALA-ASN | | Authors: | Li, H, Jiang, J. | | Deposit date: | 2018-07-13 | | Release date: | 2019-11-06 | | Last modified: | 2020-01-01 | | Method: | X-RAY DIFFRACTION (2.531 Å) | | Cite: | Targeted covalent inhibition of O-GlcNAc transferase in cells.
Chem.Commun.(Camb.), 55, 2019
|
|
6CAA
 
 | | CryoEM structure of human SLC4A4 sodium-coupled acid-base transporter NBCe1 | | Descriptor: | Electrogenic sodium bicarbonate cotransporter 1 | | Authors: | Huynh, K.W, Jiang, J, Abuladze, N, Tsirulnikov, K, Kao, L, Shao, X, Newman, D, Azimov, R, Pushkin, A, Zhou, Z.H, Kurtz, I. | | Deposit date: | 2018-01-29 | | Release date: | 2018-03-07 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | CryoEM structure of the human SLC4A4 sodium-coupled acid-base transporter NBCe1.
Nat Commun, 9, 2018
|
|
3IZX
 
 | | 3.1 Angstrom cryoEM structure of cytoplasmic polyhedrosis virus | | Descriptor: | Capsid protein VP1, Structural protein VP3, Viral structural protein 5 | | Authors: | Yu, X, Ge, P, Jiang, J, Atanasov, I, Zhou, Z.H. | | Deposit date: | 2011-01-15 | | Release date: | 2011-06-22 | | Last modified: | 2018-08-22 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Atomic Model of CPV Reveals the Mechanism Used by This Single-Shelled Virus to Economically Carry Out Functions Conserved in Multishelled Reoviruses.
Structure, 19, 2011
|
|
5IVX
 
 | | Crystal Structure of B4.2.3 T-Cell Receptor and H2-Dd P18-I10 Complex | | Descriptor: | 1,2-ETHANEDIOL, Beta-2-microglobulin, H-2 class I histocompatibility antigen, ... | | Authors: | Natarajan, K, Jiang, J, Margulies, D. | | Deposit date: | 2016-03-21 | | Release date: | 2017-03-29 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | An allosteric site in the T-cell receptor C beta domain plays a critical signalling role.
Nat Commun, 8, 2017
|
|
5IW1
 
 | |
4OI8
 
 | | RAGE is a nucleic acid receptor that promotes inflammatory responses to DNA. | | Descriptor: | 5'-D(*CP*CP*AP*TP*GP*AP*CP*TP*GP*TP*AP*GP*GP*AP*AP*AP*CP*TP*CP*TP*AP*GP*A)-3', 5'-D(*CP*TP*CP*TP*AP*GP*AP*GP*TP*TP*TP*CP*CP*TP*AP*CP*AP*GP*TP*CP*AP*TP*G)-3', Advanced glycosylation end product-specific receptor | | Authors: | Jin, T, Jiang, J, Xiao, T. | | Deposit date: | 2014-01-19 | | Release date: | 2014-04-30 | | Method: | X-RAY DIFFRACTION (3.101 Å) | | Cite: | RAGE is a nucleic acid receptor that promotes inflammatory responses to DNA.
J.Exp.Med., 210, 2013
|
|
4OI7
 
 | | RAGE recognizes nucleic acids and promotes inflammatory responses to DNA | | Descriptor: | 1,2-ETHANEDIOL, 5'-D(*CP*AP*CP*GP*TP*TP*CP*GP*TP*AP*GP*CP*AP*TP*CP*GP*TP*TP*GP*CP*AP*G)-3', 5'-D(*CP*TP*GP*CP*AP*AP*CP*GP*AP*TP*GP*CP*TP*AP*CP*GP*AP*AP*CP*GP*TP*G)-3', ... | | Authors: | Jin, T, Jiang, J, Xiao, T. | | Deposit date: | 2014-01-19 | | Release date: | 2014-04-30 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.104 Å) | | Cite: | RAGE is a nucleic acid receptor that promotes inflammatory responses to DNA.
J.Exp.Med., 210, 2013
|
|
