1QAK
 
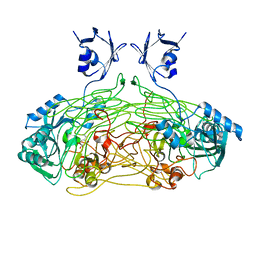 | | THE ACTIVE SITE BASE CONTROLS COFACTOR REACTIVITY IN ESCHERICHIA COLI AMINE OXIDASE : X-RAY CRYSTALLOGRAPHIC STUDIES WITH MUTATIONAL VARIANTS | | Descriptor: | CALCIUM ION, COPPER (II) ION, COPPER AMINE OXIDASE | | Authors: | Murray, J.M, Wilmot, C.M, Saysell, C.G, Jaeger, J, Knowles, P.F, Phillips, S.E, McPherson, M.J. | | Deposit date: | 1999-03-15 | | Release date: | 1999-08-24 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The active site base controls cofactor reactivity in Escherichia coli amine oxidase: x-ray crystallographic studies with mutational variants.
Biochemistry, 38, 1999
|
|
4YUK
 
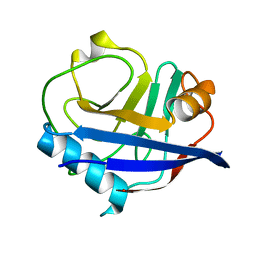 | | Multiconformer synchrotron model of CypA at 260 K | | Descriptor: | Peptidyl-prolyl cis-trans isomerase A | | Authors: | Keedy, D.A, Kenner, L.R, Warkentin, M, Woldeyes, R.A, Thompson, M.C, Brewster, A.S, Van Benschoten, A.H, Baxter, E.L, Hopkins, J.B, Uervirojnangkoorn, M, McPhillips, S.E, Song, J, Mori, R.A, Holton, J.M, Weis, W.I, Brunger, A.T, Soltis, M, Lemke, H, Gonzalez, A, Sauter, N.K, Cohen, A.E, van den Bedem, H, Thorne, R.E, Fraser, J.S. | | Deposit date: | 2015-03-18 | | Release date: | 2015-10-14 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Mapping the conformational landscape of a dynamic enzyme by multitemperature and XFEL crystallography.
Elife, 4, 2015
|
|
1QAF
 
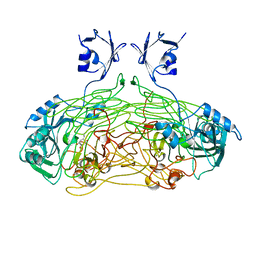 | | THE ACTIVE SITE BASE CONTROLS COFACTOR REACTIVITY IN ESCHERICHIA COLI AMINE OXIDASE : X-RAY CRYSTALLOGRAPHIC STUDIES WITH MUTATIONAL VARIANTS | | Descriptor: | CALCIUM ION, COPPER (II) ION, GLYCEROL, ... | | Authors: | Murray, J.M, Wilmot, C.M, Saysell, C.G, Jaeger, J, Knowles, P.F, Phillips, S.E, McPherson, M.J. | | Deposit date: | 1999-03-11 | | Release date: | 1999-08-23 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The active site base controls cofactor reactivity in Escherichia coli amine oxidase: x-ray crystallographic studies with mutational variants.
Biochemistry, 38, 1999
|
|
1QAL
 
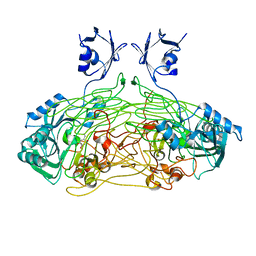 | | THE ACTIVE SITE BASE CONTROLS COFACTOR REACTIVITY IN ESCHERICHIA COLI AMINE OXIDASE : X-RAY CRYSTALLOGRAPHIC STUDIES WITH MUTATIONAL VARIANTS | | Descriptor: | CALCIUM ION, COPPER (II) ION, COPPER AMINE OXIDASE | | Authors: | Murray, J.M, Wilmot, C.M, Saysell, C.G, Jaeger, J, Knowles, P.F, Phillips, S.E, McPherson, M.J. | | Deposit date: | 1999-03-19 | | Release date: | 1999-08-24 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The active site base controls cofactor reactivity in Escherichia coli amine oxidase: x-ray crystallographic studies with mutational variants.
Biochemistry, 38, 1999
|
|
3S4G
 
 | | Low Resolution Structure of STNV complexed with RNA | | Descriptor: | Capsid protein, RNA (5'-R(P*AP*AP*A)-3'), RNA (5'-R(P*UP*UP*UP*U)-3') | | Authors: | Lane, S.W, Dennis, C.A, Lane, C.L, Trinh, C.H, Rizkallah, P.J, Stockley, P.G, Phillips, S.E.V. | | Deposit date: | 2011-05-19 | | Release date: | 2011-08-03 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (6 Å) | | Cite: | Construction and crystal structure of recombinant STNV capsids.
J.Mol.Biol., 413, 2011
|
|
4PGM
 
 | |
1AUA
 
 | | PHOSPHATIDYLINOSITOL TRANSFER PROTEIN SEC14P FROM SACCHAROMYCES CEREVISIAE | | Descriptor: | PHOSPHATIDYLINOSITOL TRANSFER PROTEIN SEC14P, octyl beta-D-glucopyranoside | | Authors: | Sha, B, Phillips, S.E, Bankaitis, V.A, Luo, M. | | Deposit date: | 1997-08-20 | | Release date: | 1997-12-24 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of the Saccharomyces cerevisiae phosphatidylinositol-transfer protein.
Nature, 391, 1998
|
|
1M0D
 
 | | Crystal Structure of Bacteriophage T7 Endonuclease I with a Wild-Type Active Site and Bound Manganese Ions | | Descriptor: | Endodeoxyribonuclease I, MANGANESE (II) ION, SULFATE ION | | Authors: | Hadden, J.M, Declais, A.C, Phillips, S.E, Lilley, D.M. | | Deposit date: | 2002-06-12 | | Release date: | 2002-07-10 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Metal ions bound at the active site of the junction-resolving enzyme T7 endonuclease I.
EMBO J., 21, 2002
|
|
1M0I
 
 | | Crystal Structure of Bacteriophage T7 Endonuclease I with a Wild-Type Active Site | | Descriptor: | SULFATE ION, endodeoxyribonuclease I | | Authors: | Hadden, J.M, Declais, A.C, Phillips, S.E, Lilley, D.M. | | Deposit date: | 2002-06-13 | | Release date: | 2002-12-18 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Metal ions bound at the active site of the junction-resolving enzyme T7 endonuclease I
Embo J., 21, 2002
|
|
2PFJ
 
 | | Crystal Structure of T7 Endo I resolvase in complex with a Holliday Junction | | Descriptor: | 27-MER, CALCIUM ION, Endodeoxyribonuclease 1 | | Authors: | Hadden, J.M, Declais, A.C, Carr, S.B, Lilley, D.M, Phillips, S.E. | | Deposit date: | 2007-04-05 | | Release date: | 2007-10-30 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | The structural basis of Holliday junction resolution by T7 endonuclease I.
Nature, 449, 2007
|
|
2VZ3
 
 | | bleached galactose oxidase | | Descriptor: | ACETATE ION, COPPER (II) ION, GALACTOSE OXIDASE | | Authors: | Rogers, M.S, Hurtado-Guerrero, R, Firbank, S.J, Halcrow, M.A, Dooley, D.M, Phillips, S.E.V, Knowles, P.F, McPherson, M.J. | | Deposit date: | 2008-07-29 | | Release date: | 2008-09-16 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Cross-Link Formation of the Cysteine 228-Tyrosine 272 Catalytic Cofactor of Galactose Oxidase Does not Require Dioxygen.
Biochemistry, 47, 2008
|
|
2W0Q
 
 | | E. coli copper amine oxidase in complex with Xenon | | Descriptor: | CALCIUM ION, COPPER (II) ION, COPPER AMINE OXIDASE, ... | | Authors: | Pirrat, P, Smith, M.A, Pearson, A.R, McPherson, M.J, Phillips, S.E.V. | | Deposit date: | 2008-08-20 | | Release date: | 2008-12-16 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Structure of a Xenon Derivative of Escherichia Coli Copper Amine Oxidase: Confirmation of the Proposed Oxygen-Entry Pathway.
Acta Crystallogr.,Sect.F, 64, 2008
|
|
1T2X
 
 | | Glactose oxidase C383S mutant identified by directed evolution | | Descriptor: | ACETATE ION, COPPER (II) ION, Galactose Oxidase, ... | | Authors: | Wilkinson, D, Akumanyi, N, Hurtado-Guerrero, R, Dawkes, H, Knowles, P.F, Phillips, S.E.V, McPherson, M.J. | | Deposit date: | 2004-04-23 | | Release date: | 2004-05-18 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural and kinetic studies of a series of mutants of galactose oxidase identified by directed evolution.
Protein Eng.Des.Sel., 17, 2004
|
|
4UUI
 
 | |
6GQW
 
 | | KRAS-169 Q61H GPPNHP + CH-1 | | Descriptor: | GTPase KRas, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER, ... | | Authors: | Cruz-Migoni, A, Quevedo, C.E, Carr, S.B, Phillips, S.E.V, Rabbitts, T.H. | | Deposit date: | 2018-06-08 | | Release date: | 2019-02-06 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure-based development of new RAS-effector inhibitors from a combination of active and inactive RAS-binding compounds.
Proc. Natl. Acad. Sci. U.S.A., 116, 2019
|
|
6GOM
 
 | | KRAS-169 Q61H GPPNHP + PPIN-1 | | Descriptor: | (6~{S})-1-(1~{H}-imidazol-4-ylcarbonyl)-6-[(4-phenylphenyl)methyl]-4-propyl-1,4-diazepan-5-one, CITRIC ACID, GTPase KRas, ... | | Authors: | Cruz-Migoni, A, Canning, P, Quevedo, C.E, Carr, S.B, Phillips, S.E.V, Rabbitts, T.H. | | Deposit date: | 2018-06-01 | | Release date: | 2019-02-06 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Structure-based development of new RAS-effector inhibitors from a combination of active and inactive RAS-binding compounds.
Proc. Natl. Acad. Sci. U.S.A., 116, 2019
|
|
6GQT
 
 | | KRAS-169 Q61H GPPNHP + PPIN-2 | | Descriptor: | CITRIC ACID, GTPase KRas, MAGNESIUM ION, ... | | Authors: | Cruz-Migoni, A, Canning, P, Quevedo, C.E, Carr, S.B, Phillips, S.E.V, Rabbitts, T.H. | | Deposit date: | 2018-06-08 | | Release date: | 2019-02-06 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Structure-based development of new RAS-effector inhibitors from a combination of active and inactive RAS-binding compounds.
Proc. Natl. Acad. Sci. U.S.A., 116, 2019
|
|
6GQX
 
 | | KRAS-169 Q61H GPPNHP + CH-2 | | Descriptor: | GTPase KRas, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER, ... | | Authors: | Cruz-Migoni, A, Quevedo, C.E, Carr, S.B, Phillips, S.E.V, Rabbitts, T.H. | | Deposit date: | 2018-06-08 | | Release date: | 2019-02-06 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure-based development of new RAS-effector inhibitors from a combination of active and inactive RAS-binding compounds.
Proc. Natl. Acad. Sci. U.S.A., 116, 2019
|
|
5LMM
 
 | | Structure of E coli Hydrogenase Hyd-1 mutant E28Q | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CARBONMONOXIDE-(DICYANO) IRON, CHLORIDE ION, ... | | Authors: | Carr, S.B, Phillips, S.E.V, Evans, R.M, Brooke, E.J, Armstrong, F.A. | | Deposit date: | 2016-08-01 | | Release date: | 2017-08-16 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Kinetic consequences of re-engineering the outer shell "canopy" above the active site of a [NiFe]-hydrogenase.
To Be Published
|
|
5MVL
 
 | | Crystal structure of an A-DNA dodecamer containing 5-bromouracil | | Descriptor: | Brominated DNA dodecamer, MAGNESIUM ION | | Authors: | Hardwick, J.S, Ptchelkine, D, Phillips, S.E.V, Brown, T. | | Deposit date: | 2017-01-16 | | Release date: | 2017-05-10 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.405 Å) | | Cite: | 5-Formylcytosine does not change the global structure of DNA.
Nat. Struct. Mol. Biol., 24, 2017
|
|
5MVT
 
 | | Crystal structure of an A-DNA dodecamer featuring an alternating pyrimidine-purine sequence | | Descriptor: | COBALT (III) ION, DNA | | Authors: | Hardwick, J.S, Ptchelkine, D, Phillips, S.E.V, Brown, T. | | Deposit date: | 2017-01-17 | | Release date: | 2017-05-10 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.896 Å) | | Cite: | 5-Formylcytosine does not change the global structure of DNA.
Nat. Struct. Mol. Biol., 24, 2017
|
|
5MVQ
 
 | | Crystal structure of an unmodified, self-complementary dodecamer. | | Descriptor: | DNA, MAGNESIUM ION | | Authors: | Hardwick, J.S, Ptchelkine, D, Phillips, S.E.V, Brown, T. | | Deposit date: | 2017-01-17 | | Release date: | 2017-05-10 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.604 Å) | | Cite: | 5-Formylcytosine does not change the global structure of DNA.
Nat. Struct. Mol. Biol., 24, 2017
|
|
5MVU
 
 | |
5MVP
 
 | | Crystal structure of an A-DNA dodecamer containing the GGGCCC motif | | Descriptor: | DNA, POTASSIUM ION | | Authors: | Hardwick, J.S, Ptchelkine, D, Phillips, S.E.V, Brown, T. | | Deposit date: | 2017-01-17 | | Release date: | 2017-05-10 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.606 Å) | | Cite: | 5-Formylcytosine does not change the global structure of DNA.
Nat. Struct. Mol. Biol., 24, 2017
|
|
5MVK
 
 | |
