1X1L
 
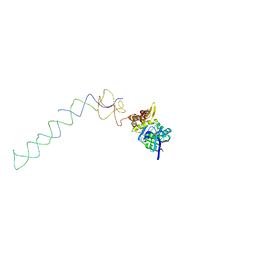 | | Interaction of ERA,a GTPase protein, with the 3'minor domain of the 16S rRNA within the THERMUS THERMOPHILUS 30S subunit. | | Descriptor: | GTP-binding protein era, RNA (130-MER) | | Authors: | Sharma, M.R, Barat, C, Agrawal, R.K. | | Deposit date: | 2005-04-06 | | Release date: | 2005-05-17 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (13.5 Å) | | Cite: | Interaction of Era with the 30S Ribosomal Subunit Implications for 30S Subunit Assembly
Mol.Cell, 18, 2005
|
|
8BHD
 
 | | N-terminal domain of Plasmodium berghei glutamyl-tRNA synthetase (Tbxo4 derivative crystal structure) | | Descriptor: | GLYCEROL, Glutamate--tRNA ligase, SULFATE ION, ... | | Authors: | Benas, P, Jaramillo Ponce, J.R, Legrand, P, Frugier, M, Sauter, C. | | Deposit date: | 2022-10-31 | | Release date: | 2023-01-25 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (3.17 Å) | | Cite: | Solution X-ray scattering highlights discrepancies in Plasmodium multi-aminoacyl-tRNA synthetase complexes.
Protein Sci., 32, 2023
|
|
8SKQ
 
 | |
1E1L
 
 | |
1E1K
 
 | |
1E1M
 
 | |
1E1N
 
 | |
3SYU
 
 | | Re-refined coordinates for pdb entry 1det - ribonuclease T1 carboxymethylated at GLU 58 in complex with 2'GMP | | Descriptor: | GUANOSINE-2'-MONOPHOSPHATE, Guanyl-specific ribonuclease T1, SODIUM ION, ... | | Authors: | Smart, O.S, Womack, T.O, Bricogne, G. | | Deposit date: | 2011-07-18 | | Release date: | 2012-03-28 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Exploiting structure similarity in refinement: automated NCS and target-structure restraints in BUSTER.
Acta Crystallogr.,Sect.D, 68, 2012
|
|
7BBM
 
 | | Mutant nitrobindin M75L/H76L/Q96C/M148L (NB4H) from Arabidopsis thaliana with cofactor MnPPIX | | Descriptor: | 1,2-ETHANEDIOL, MANGANESE PROTOPORPHYRIN IX, UPF0678 fatty acid-binding protein-like protein At1g79260 | | Authors: | Minges, A, Sauer, D.F, Wittwer, M, Markel, U, Spiertz, M, Schiffels, J, Davari, M.D, Okuda, J, Schwaneberg, U, Groth, G. | | Deposit date: | 2020-12-18 | | Release date: | 2021-05-26 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.14 Å) | | Cite: | Chemogenetic engineering of nitrobindin toward an artificial epoxygenase
Catalysis Science And Technology, 2021
|
|
5TZR
 
 | | GPR40 in complex with partial agonist MK-8666 | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, (5aR,6S,6aS)-3-({2',6'-dimethyl-4'-[3-(methylsulfonyl)propoxy][1,1'-biphenyl]-3-yl}methoxy)-5,5a,6,6a-tetrahydrocyclopropa[4,5]cyclopenta[1,2-c]pyridine-6-carboxylic acid, Free fatty acid receptor 1,Endolysin,Free fatty acid receptor 1, ... | | Authors: | Lu, J, Byrne, N, Patel, S, Sharma, S, Soisson, S.M. | | Deposit date: | 2016-11-22 | | Release date: | 2017-06-07 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis for the cooperative allosteric activation of the free fatty acid receptor GPR40.
Nat. Struct. Mol. Biol., 24, 2017
|
|
8A67
 
 | |
5TZY
 
 | | GPR40 in complex with AgoPAM AP8 and partial agonist MK-8666 | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, (2S,3R)-3-cyclopropyl-3-[(2R)-2-(1-{(1S)-1-[5-fluoro-2-(trifluoromethoxy)phenyl]ethyl}piperidin-4-yl)-3,4-dihydro-2H-1-benzopyran-7-yl]-2-methylpropanoic acid, (5aR,6S,6aS)-3-({2',6'-dimethyl-4'-[3-(methylsulfonyl)propoxy][1,1'-biphenyl]-3-yl}methoxy)-5,5a,6,6a-tetrahydrocyclopropa[4,5]cyclopenta[1,2-c]pyridine-6-carboxylic acid, ... | | Authors: | Lu, J, Byrne, N, Patel, S, Sharma, S, Soisson, S.M. | | Deposit date: | 2016-11-22 | | Release date: | 2017-06-07 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.22 Å) | | Cite: | Structural basis for the cooperative allosteric activation of the free fatty acid receptor GPR40.
Nat. Struct. Mol. Biol., 24, 2017
|
|
3N43
 
 | | Crystal structures of the mature envelope glycoprotein complex (trypsin cleavage) of Chikungunya virus. | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, ... | | Authors: | Voss, J, Vaney, M.C, Duquerroy, S, Rey, F.A. | | Deposit date: | 2010-05-21 | | Release date: | 2010-12-01 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.58 Å) | | Cite: | Glycoprotein organization of Chikungunya virus particles revealed by X-ray crystallography.
Nature, 468, 2010
|
|
3N40
 
 | | Crystal structure of the immature envelope glycoprotein complex of Chikungunya virus. | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Voss, J, Vaney, M.C, Duquerroy, S, Rey, F.A. | | Deposit date: | 2010-05-21 | | Release date: | 2010-12-01 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Glycoprotein organization of Chikungunya virus particles revealed by X-ray crystallography.
Nature, 468, 2010
|
|
3N42
 
 | | Crystal structures of the mature envelope glycoprotein complex (furin cleavage) of Chikungunya virus. | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, E1 envelope glycoprotein, ... | | Authors: | Voss, J, Vaney, M.C, Duquerroy, S, Rey, F.A. | | Deposit date: | 2010-05-21 | | Release date: | 2010-12-01 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Glycoprotein organization of Chikungunya virus particles revealed by X-ray crystallography.
Nature, 468, 2010
|
|
3N44
 
 | | Crystal structure of the mature envelope glycoprotein complex (trypsin cleavage; Osmium soak) of Chikungunya virus. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, E1 envelope glycoprotein, ... | | Authors: | Voss, J, Vaney, M.C, Duquerroy, S, Rey, F.A. | | Deposit date: | 2010-05-21 | | Release date: | 2010-12-01 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Glycoprotein organization of Chikungunya virus particles revealed by X-ray crystallography.
Nature, 468, 2010
|
|
3N41
 
 | | Crystal structure of the mature envelope glycoprotein complex (spontaneous cleavage) of Chikungunya virus. | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, E1 envelope glycoprotein, ... | | Authors: | Voss, J, Vaney, M.C, Duquerroy, S, Rey, F.A. | | Deposit date: | 2010-05-21 | | Release date: | 2010-12-01 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.01 Å) | | Cite: | Glycoprotein organization of Chikungunya virus particles revealed by X-ray crystallography.
Nature, 468, 2010
|
|
6RTD
 
 | | Dihydro-heme d1 dehydrogenase NirN in complex with DHE | | Descriptor: | (R,R)-2,3-BUTANEDIOL, Cytochrome c, HEME C, ... | | Authors: | Kluenemann, T, Preuss, A, Layer, G, Blankenfeldt, W. | | Deposit date: | 2019-05-23 | | Release date: | 2019-06-19 | | Last modified: | 2019-08-28 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | Crystal Structure of Dihydro-Heme d1Dehydrogenase NirN from Pseudomonas aeruginosa Reveals Amino Acid Residues Essential for Catalysis.
J.Mol.Biol., 431, 2019
|
|
6RTE
 
 | | Dihydro-heme d1 dehydrogenase NirN in complex with DHE | | Descriptor: | (R,R)-2,3-BUTANEDIOL, Cytochrome c, HEME C | | Authors: | Kluenemann, T, Preuss, A, Layer, G, Blankenfeldt, W. | | Deposit date: | 2019-05-23 | | Release date: | 2019-06-19 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Crystal Structure of Dihydro-Heme d1Dehydrogenase NirN from Pseudomonas aeruginosa Reveals Amino Acid Residues Essential for Catalysis.
J.Mol.Biol., 431, 2019
|
|
7BHE
 
 | | DARPin_D5/Her3 domain 4 complex, monoclinic crystals | | Descriptor: | ACETATE ION, DARPin_D5, GLYCEROL, ... | | Authors: | Mittl, P.R.E, Radom, F, Pluckthun, A. | | Deposit date: | 2021-01-11 | | Release date: | 2021-11-24 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.297 Å) | | Cite: | Crystal structures of HER3 extracellular domain 4 in complex with the designed ankyrin-repeat protein D5.
Acta Crystallogr.,Sect.F, 77, 2021
|
|
7BHF
 
 | | DARPin_D5/Her3 domain 4 complex, orthorhombic crystals | | Descriptor: | ACETATE ION, DARPin_D5, Isoform 4 of Receptor tyrosine-protein kinase erbB-3 | | Authors: | Mittl, P.R.E, Radom, F, Pluckthun, A. | | Deposit date: | 2021-01-11 | | Release date: | 2021-11-24 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.997 Å) | | Cite: | Crystal structures of HER3 extracellular domain 4 in complex with the designed ankyrin-repeat protein D5.
Acta Crystallogr.,Sect.F, 77, 2021
|
|
2BK9
 
 | | Drosophila Melanogaster globin | | Descriptor: | 3-CYCLOHEXYL-1-PROPYLSULFONIC ACID, CG9734-PA, CHLORIDE ION, ... | | Authors: | de Sanctis, D, Dewilde, S, Pesce, A, Moens, L, Ascenzi, P, Hankeln, T, Burmester, T, Ponassi, M, Nardini, M, Bolognesi, M. | | Deposit date: | 2005-02-14 | | Release date: | 2005-05-20 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Bishistidyl Heme Hexacoordination, a Key Structural Property in Drosophila Melanogaster Hemoglobin
J.Biol.Chem., 280, 2005
|
|
1YDI
 
 | |
7ZFM
 
 | | Engineered Protein Targeting the Zika Viral Envelope Fusion Loop | | Descriptor: | 1,2-ETHANEDIOL, ACETIC ACID, HEXAETHYLENE GLYCOL, ... | | Authors: | Athayde, D, Archer, M, Viana, I.F.T, Adan, W.C.S, Xavier, L.S.S, Lins, R.D. | | Deposit date: | 2022-04-01 | | Release date: | 2022-08-17 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.711 Å) | | Cite: | In Vitro Neutralisation of Zika Virus by an Engineered Protein Targeting the Viral Envelope Fusion Loop
SSRN, 2022
|
|
1I6M
 
 | |
