4HOW
 
 | | The crystal structure of isomaltulose synthase from Erwinia rhapontici NX5 | | Descriptor: | CALCIUM ION, GLYCEROL, Sucrose isomerase | | Authors: | Xu, Z, Li, S, Xu, H, Zhou, J. | | Deposit date: | 2012-10-22 | | Release date: | 2013-10-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The Structural Basis of Erwinia rhapontici Isomaltulose Synthase
Plos One, 8, 2013
|
|
4HPH
 
 | | The crystal structure of isomaltulose synthase mutant E295Q from Erwinia rhapontici NX5 in complex with its natural substrate sucrose | | Descriptor: | CALCIUM ION, GLYCEROL, Sucrose isomerase, ... | | Authors: | Xu, Z, Li, S, Xu, H, Zhou, J. | | Deposit date: | 2012-10-23 | | Release date: | 2013-10-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The Structural Basis of Erwinia rhapontici Isomaltulose Synthase
Plos One, 8, 2013
|
|
5C71
 
 | | The structure of Aspergillus oryzae a-glucuronidase complexed with glycyrrhetinic acid monoglucuronide | | Descriptor: | (3BETA,5BETA,14BETA)-3-HYDROXY-11-OXOOLEAN-12-EN-29-OIC ACID, Glucuronidase, alpha-D-glucopyranuronic acid | | Authors: | Sun, H.L, Lv, B, Huang, S, Li, C, Jiang, T. | | Deposit date: | 2015-06-24 | | Release date: | 2016-06-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.62 Å) | | Cite: | Structure-guided engineering of the substrate specificity of a fungal beta-glucuronidase toward triterpenoid saponins.
J.Biol.Chem., 293, 2018
|
|
8G2U
 
 | | Time-resolved cryo-EM study of the 70S recycling by the HflX:control-apo-70S at 900ms | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Bhattacharjee, S, Brown, P.Z, Frank, J. | | Deposit date: | 2023-02-06 | | Release date: | 2023-12-06 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Time resolution in cryo-EM using a PDMS-based microfluidic chip assembly and its application to the study of HflX-mediated ribosome recycling.
Cell, 187, 2024
|
|
8G34
 
 | | Time-resolved cryo-EM study of the 70S recycling by the HflX:1st intermediate | | Descriptor: | 16S, 23S, 30S ribosomal protein S10, ... | | Authors: | Bhattacharjee, S, Brown, P.Z, Frank, J. | | Deposit date: | 2023-02-06 | | Release date: | 2023-12-06 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Time resolution in cryo-EM using a PDMS-based microfluidic chip assembly and its application to the study of HflX-mediated ribosome recycling.
Cell, 187, 2024
|
|
8G38
 
 | | Time-resolved cryo-EM study of the 70S recycling by the HflX:3rd Intermediate | | Descriptor: | 16S, 23S, 30S ribosomal protein S10, ... | | Authors: | Bhattacharjee, S, Brown, P.Z, Frank, J. | | Deposit date: | 2023-02-07 | | Release date: | 2023-12-06 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Time resolution in cryo-EM using a PDMS-based microfluidic chip assembly and its application to the study of HflX-mediated ribosome recycling.
Cell, 187, 2024
|
|
8G31
 
 | | Time-resolved cryo-EM study of the 70S recycling by the HflX:2nd Intermediate | | Descriptor: | 16S, 23S, 30S ribosomal protein S10, ... | | Authors: | Bhattacharjee, S, Brown, P.Z, Frank, J. | | Deposit date: | 2023-02-06 | | Release date: | 2023-12-06 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Time resolution in cryo-EM using a PDMS-based microfluidic chip assembly and its application to the study of HflX-mediated ribosome recycling.
Cell, 187, 2024
|
|
4HOZ
 
 | | The crystal structure of isomaltulose synthase mutant D241A from Erwinia rhapontici NX5 in complex with D-glucose | | Descriptor: | CALCIUM ION, GLYCEROL, Sucrose isomerase, ... | | Authors: | Xu, Z, Li, S, Xu, H, Zhou, J. | | Deposit date: | 2012-10-23 | | Release date: | 2013-10-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The Structural Basis of Erwinia rhapontici Isomaltulose Synthase
Plos One, 8, 2013
|
|
3UE1
 
 | | Crystal strucuture of Acinetobacter baumanni PBP1A in complex with MC-1 | | Descriptor: | (4S,7Z)-7-(2-amino-1,3-thiazol-4-yl)-1-[({4-[(2R)-2,3-dihydroxypropyl]-3-(4,5-dihydroxypyridin-2-yl)-5-oxo-4,5-dihydro- 1H-1,2,4-triazol-1-yl}sulfonyl)amino]-4-formyl-10,10-dimethyl-1,6-dioxo-9-oxa-2,5,8-triazaundec-7-en-11-oate, Penicillin-binding protein 1a | | Authors: | Han, S. | | Deposit date: | 2011-10-28 | | Release date: | 2011-12-14 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.73 Å) | | Cite: | Distinctive attributes of beta-lactam target proteins in Acinetobacter baumannii relevant to development of new antibiotics
J.Am.Chem.Soc., 133, 2011
|
|
3UE3
 
 | | Crystal structure of Acinetobacter baumanni PBP3 | | Descriptor: | Septum formation, penicillin binding protein 3, peptidoglycan synthetase | | Authors: | Han, S. | | Deposit date: | 2011-10-28 | | Release date: | 2011-12-14 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Distinctive attributes of beta-lactam target proteins in Acinetobacter baumannii relevant to development of new antibiotics
J.Am.Chem.Soc., 133, 2011
|
|
3VKD
 
 | | Crystal structure of MoeO5 soaked with 3-phosphoglycerate | | Descriptor: | (2R)-3-(phosphonooxy)-2-{[(2Z,6E)-3,7,11-trimethyldodeca-2,6,10-trien-1-yl]oxy}propanoic acid, MAGNESIUM ION, MoeO5, ... | | Authors: | Ren, F, Ko, T.-P, Huang, C.-H, Guo, R.-T. | | Deposit date: | 2011-11-12 | | Release date: | 2012-05-09 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Insights into the mechanism of the antibiotic-synthesizing enzyme MoeO5 from crystal structures of different complexes
Angew.Chem.Int.Ed.Engl., 51, 2012
|
|
3VK5
 
 | | Crystal structure of MoeO5 in complex with its product FPG | | Descriptor: | (2R)-3-(phosphonooxy)-2-{[(2Z,6E)-3,7,11-trimethyldodeca-2,6,10-trien-1-yl]oxy}propanoic acid, MAGNESIUM ION, MoeO5 | | Authors: | Ren, F, Ko, T.-P, Huang, C.-H, Guo, R.-T. | | Deposit date: | 2011-11-08 | | Release date: | 2012-05-09 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.39 Å) | | Cite: | Insights into the mechanism of the antibiotic-synthesizing enzyme MoeO5 from crystal structures of different complexes
Angew.Chem.Int.Ed.Engl., 51, 2012
|
|
3VKC
 
 | | Crystal structure of MoeO5 soaked with pyrophosphate | | Descriptor: | (2R)-3-(phosphonooxy)-2-{[(2Z,6E)-3,7,11-trimethyldodeca-2,6,10-trien-1-yl]oxy}propanoic acid, MAGNESIUM ION, MoeO5, ... | | Authors: | Ren, F, Ko, T.-P, Huang, C.-H, Guo, R.-T. | | Deposit date: | 2011-11-12 | | Release date: | 2012-05-09 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Insights into the mechanism of the antibiotic-synthesizing enzyme MoeO5 from crystal structures of different complexes
Angew.Chem.Int.Ed.Engl., 51, 2012
|
|
3VKB
 
 | | Crystal structure of MoeO5 soaked with FsPP overnight | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, MoeO5, ... | | Authors: | Ren, F, Ko, T.-P, Huang, C.-H, Guo, R.-T. | | Deposit date: | 2011-11-11 | | Release date: | 2012-05-09 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Insights into the mechanism of the antibiotic-synthesizing enzyme MoeO5 from crystal structures of different complexes
Angew.Chem.Int.Ed.Engl., 51, 2012
|
|
3VKA
 
 | | Crystal structure of MoeO5 soaked for 3 hours in FsPP | | Descriptor: | (2R)-3-(phosphonooxy)-2-{[(2Z,6E)-3,7,11-trimethyldodeca-2,6,10-trien-1-yl]oxy}propanoic acid, MAGNESIUM ION, MoeO5 | | Authors: | Ren, F, Ko, T.-P, Huang, C.-H, Guo, R.-T. | | Deposit date: | 2011-11-11 | | Release date: | 2012-05-09 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Insights into the mechanism of the antibiotic-synthesizing enzyme MoeO5 from crystal structures of different complexes
Angew.Chem.Int.Ed.Engl., 51, 2012
|
|
3UDI
 
 | |
3UDF
 
 | | Crystal structure of Apo PBP1a from Acinetobacter baumannii | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Penicillin-binding protein 1a | | Authors: | Han, S. | | Deposit date: | 2011-10-28 | | Release date: | 2011-12-14 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Distinctive attributes of beta-lactam target proteins in Acinetobacter baumannii relevant to development of new antibiotics
J.Am.Chem.Soc., 133, 2011
|
|
3UE0
 
 | | Crystal structure of Acinetobacter baumannii PBP1a in complex with Aztreonam | | Descriptor: | 2-({[(1Z)-1-(2-amino-1,3-thiazol-4-yl)-2-oxo-2-{[(2S,3S)-1-oxo-3-(sulfoamino)butan-2-yl]amino}ethylidene]amino}oxy)-2-methylpropanoic acid, Penicillin-binding protein 1a | | Authors: | Han, S. | | Deposit date: | 2011-10-28 | | Release date: | 2011-12-14 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Distinctive attributes of beta-lactam target proteins in Acinetobacter baumannii relevant to development of new antibiotics
J.Am.Chem.Soc., 133, 2011
|
|
3UDX
 
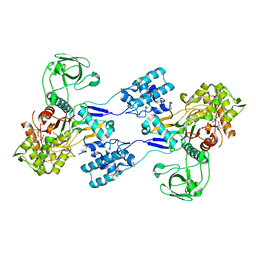 | | Crystal structure of Acinetobacter baumannii PBP1a in complex with Imipenem | | Descriptor: | (5R)-5-[(1S,2R)-1-formyl-2-hydroxypropyl]-3-[(2-{[(E)-iminomethyl]amino}ethyl)sulfanyl]-4,5-dihydro-1H-pyrrole-2-carbox ylic acid, Penicillin-binding protein 1a | | Authors: | Han, S. | | Deposit date: | 2011-10-28 | | Release date: | 2011-12-14 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Distinctive attributes of beta-lactam target proteins in Acinetobacter baumannii relevant to development of new antibiotics
J.Am.Chem.Soc., 133, 2011
|
|
5V8U
 
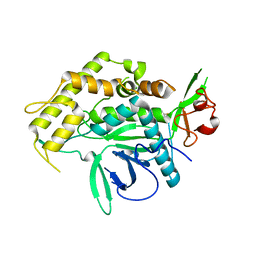 | |
5V8R
 
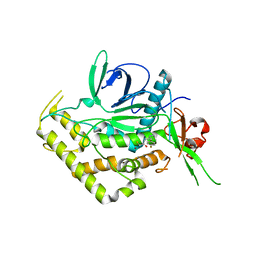 | |
2N7E
 
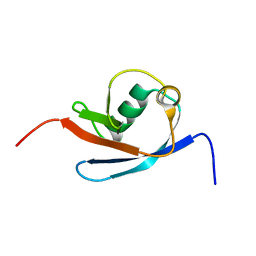 | |
5V8P
 
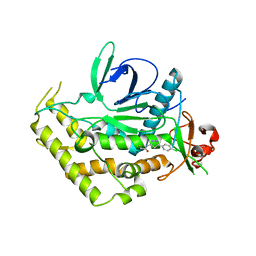 | |
6WB9
 
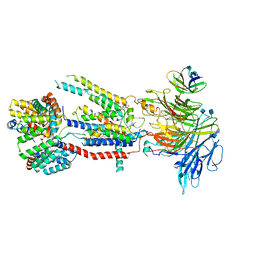 | | Structure of the S. cerevisiae ER membrane complex | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, 2-acetamido-2-deoxy-beta-D-glucopyranose, ER membrane protein complex subunit 1, ... | | Authors: | Bai, L, Li, H. | | Deposit date: | 2020-03-26 | | Release date: | 2020-06-03 | | Last modified: | 2020-09-02 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structure of the ER membrane complex, a transmembrane-domain insertase.
Nature, 584, 2020
|
|
6W8I
 
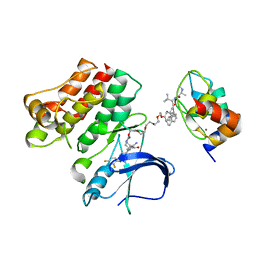 | | Ternary complex structure - BTK cIAP compound 15 | | Descriptor: | 14-{[(3S)-2-(N-methyl-L-alanyl-3-methyl-L-valyl)-3-{[(1R)-1,2,3,4-tetrahydronaphthalen-1-yl]carbamoyl}-1,2,3,4-tetrahydroisoquinolin-7-yl]oxy}-3,6,9,12-tetraoxatetradecan-1-yl (3R)-3-{5-amino-4-carbamoyl-3-[4-(2,4-difluorophenoxy)phenyl]-1H-pyrazol-1-yl}piperidine-1-carboxylate, Baculoviral IAP repeat-containing protein 2, Tyrosine-protein kinase BTK, ... | | Authors: | Calabrese, M.F, Schiemer, J.S. | | Deposit date: | 2020-03-20 | | Release date: | 2020-11-18 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | Structural Characterization of BTK:PROTAC:cIAP Ternary Complexes: From Snapshots to Ensembles
Nat.Chem.Biol., 2020
|
|
