6O3O
 
 | | Structure of human DNAM-1 (CD226) bound to nectin-like protein-5 (necl-5) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CD226 antigen, ... | | Authors: | Deuss, F.A, Watson, G.M, Rossjohn, J, Berry, R. | | Deposit date: | 2019-02-27 | | Release date: | 2019-07-10 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural basis for the recognition of nectin-like protein-5 by the human-activating immune receptor, DNAM-1.
J.Biol.Chem., 294, 2019
|
|
6MYL
 
 | | The Prp8 intein-cisplatin complex | | Descriptor: | PLATINUM (II) ION, Pre-mRNA-processing-splicing factor 8 | | Authors: | Li, Z, Li, H. | | Deposit date: | 2018-11-01 | | Release date: | 2019-11-06 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (3.06 Å) | | Cite: | Cisplatin protects mice from challenge ofCryptococcus neoformansby targeting the Prp8 intein.
Emerg Microbes Infect, 8, 2019
|
|
8THN
 
 | | KcsA M96V mutant with Y78ester in High K+ | | Descriptor: | KcsA Fab Heavy Chain, KcsA Fab Light Chain, POTASSIUM ION, ... | | Authors: | Reddi, R, Valiyaveetil, F.I. | | Deposit date: | 2023-07-17 | | Release date: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | A facile approach for incorporating tyrosine esters to probe ion-binding sites and backbone hydrogen bonds.
J.Biol.Chem., 300, 2023
|
|
7SN4
 
 | |
7SQD
 
 | |
7SN9
 
 | |
6FCM
 
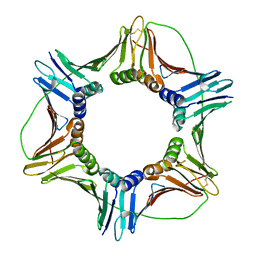 | | Crystal structure of human PCNA | | Descriptor: | Proliferating cell nuclear antigen | | Authors: | Housset, D, Frachet, P. | | Deposit date: | 2017-12-20 | | Release date: | 2019-01-30 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Cytosolic PCNA interacts with p47phox and controls NADPH oxidase NOX2 activation in neutrophils.
J.Exp.Med., 216, 2019
|
|
6FCN
 
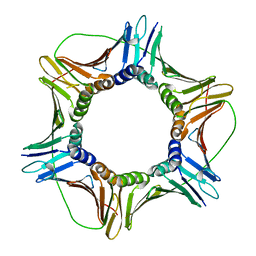 | |
8CXM
 
 | | Cryo-EM structure of the supercoiled E. coli K12 flagellar filament core, Normal waveform | | Descriptor: | Flagellin | | Authors: | Sonani, R.R, Kreutzberger, M.A.B, Sebastian, A.L, Scharf, B, Egelman, E.H. | | Deposit date: | 2022-05-21 | | Release date: | 2022-09-07 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.21 Å) | | Cite: | Convergent evolution in the supercoiling of prokaryotic flagellar filaments.
Cell, 185, 2022
|
|
5UZG
 
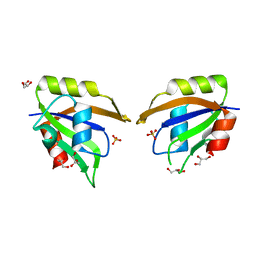 | | Crystal structure of Glorund qRRM1 domain | | Descriptor: | AT27789p, GLYCEROL, SULFATE ION | | Authors: | Teramoto, T, Hall, T.M.T. | | Deposit date: | 2017-02-26 | | Release date: | 2017-03-22 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.541 Å) | | Cite: | The Drosophila hnRNP F/H Homolog Glorund Uses Two Distinct RNA-Binding Modes to Diversify Target Recognition.
Cell Rep, 19, 2017
|
|
8DHR
 
 | | An ester mutant of SfGFP | | Descriptor: | Green fluorescent protein | | Authors: | Reddi, R, Valiyaveetil, F.I. | | Deposit date: | 2022-06-28 | | Release date: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | A facile approach for incorporating tyrosine esters to probe ion-binding sites and backbone hydrogen bonds.
J.Biol.Chem., 300, 2023
|
|
5UZM
 
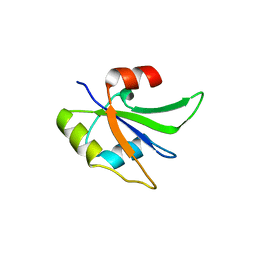 | | Crystal structure of Glorund qRRM2 domain | | Descriptor: | AT27789p | | Authors: | Teramoto, T, Hall, T.M.T. | | Deposit date: | 2017-02-27 | | Release date: | 2017-03-22 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.552 Å) | | Cite: | The Drosophila hnRNP F/H Homolog Glorund Uses Two Distinct RNA-Binding Modes to Diversify Target Recognition.
Cell Rep, 19, 2017
|
|
5UZN
 
 | | Crystal structure of Glorund qRRM3 domain | | Descriptor: | AT27789p, GLYCEROL, SULFATE ION | | Authors: | Teramoto, T, Hall, T.M.T. | | Deposit date: | 2017-02-27 | | Release date: | 2017-03-29 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | The Drosophila hnRNP F/H Homolog Glorund Uses Two Distinct RNA-Binding Modes to Diversify Target Recognition.
Cell Rep, 19, 2017
|
|
2LQ1
 
 | | Solution structure of de novo designed antifreeze peptide 3 | | Descriptor: | de novo designed antifreeze peptide 3 | | Authors: | Bhunia, A. | | Deposit date: | 2012-02-21 | | Release date: | 2012-10-24 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution structures, dynamics, and ice growth inhibitory activity of Peptide fragments derived from an antarctic yeast protein
Plos One, 7, 2012
|
|
2LQ0
 
 | | Solution structure of de novo designed antifreeze peptide 1m | | Descriptor: | de novo designed antifreeze peptide 1m | | Authors: | Bhunia, A. | | Deposit date: | 2012-02-21 | | Release date: | 2012-10-24 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution structures, dynamics, and ice growth inhibitory activity of Peptide fragments derived from an antarctic yeast protein
Plos One, 7, 2012
|
|
2LQ2
 
 | | Solution structure of de novo designed peptide 4m | | Descriptor: | de novo designed antifreeze peptide 4m | | Authors: | Bhunia, A. | | Deposit date: | 2012-02-22 | | Release date: | 2012-10-24 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution structures, dynamics, and ice growth inhibitory activity of Peptide fragments derived from an antarctic yeast protein
Plos One, 7, 2012
|
|
