5WOP
 
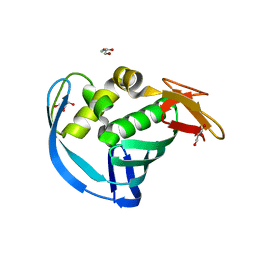 | | High Resolution Structure of Mutant CA09-PB2cap | | Descriptor: | GLYCEROL, Polymerase PB2 | | Authors: | Constantinides, A.E, Gumpper, R.H, Severin, C, Luo, M. | | Deposit date: | 2017-08-02 | | Release date: | 2017-12-20 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | High-resolution structure of the Influenza A virus PB2cap binding domain illuminates the changes induced by ligand binding.
Acta Crystallogr F Struct Biol Commun, 74, 2018
|
|
1T0Y
 
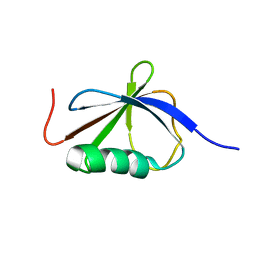 | | Solution Structure of a Ubiquitin-Like Domain from Tubulin-binding Cofactor B | | Descriptor: | tubulin folding cofactor B | | Authors: | Lytle, B.L, Peterson, F.C, Qui, S.H, Luo, M, Volkman, B.F, Markley, J.L, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2004-04-13 | | Release date: | 2004-04-27 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of a Ubiquitin-like Domain from Tubulin-binding Cofactor B.
J.Biol.Chem., 279, 2004
|
|
5EG7
 
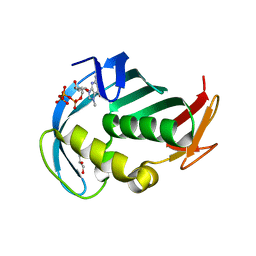 | | The cap binding site of influenza virus protein PB2 as a drug target | | Descriptor: | 7N-METHYL-8-HYDROGUANOSINE-5'-TRIPHOSPHATE, GLYCEROL, Polymerase basic protein 2 | | Authors: | Severin, C, Rocha de Moura, T, Liu, Y, Li, K, Zheng, X, Luo, M. | | Deposit date: | 2015-10-26 | | Release date: | 2016-02-10 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | The cap-binding site of influenza virus protein PB2 as a drug target.
Acta Crystallogr D Struct Biol, 72, 2016
|
|
5EG8
 
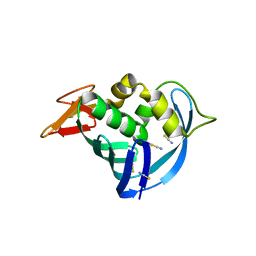 | | The cap binding site of influenza virus protein PB2 as a drug target | | Descriptor: | Polymerase basic protein 2, THIOCYANATE ION | | Authors: | Liu, Y, Zheng, X, Severin, C, Rocha de Moura, T, Li, K, Luo, M. | | Deposit date: | 2015-10-26 | | Release date: | 2016-02-10 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | The cap-binding site of influenza virus protein PB2 as a drug target.
Acta Crystallogr D Struct Biol, 72, 2016
|
|
5EG9
 
 | | The cap binding site of influenza virus protein PB2 as a drug target | | Descriptor: | Polymerase basic protein 2 | | Authors: | Severin, C, Rocha de Moura, T, Liu, Y, Li, K, Zheng, X, Luo, M. | | Deposit date: | 2015-10-26 | | Release date: | 2016-02-10 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The cap-binding site of influenza virus protein PB2 as a drug target.
Acta Crystallogr D Struct Biol, 72, 2016
|
|
6M4P
 
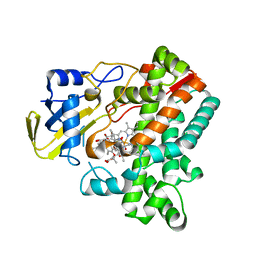 | | Cytochrome P450 monooxygenase StvP2 substrate-bound structure | | Descriptor: | 6-methoxy-streptovaricin C, Cytochrome P450, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Sun, G, Hu, C, Mei, Q, Luo, M, Chen, X, Li, Z, Liu, Y, Deng, Z, Zhang, Z, Sun, Y. | | Deposit date: | 2020-03-08 | | Release date: | 2020-08-12 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Uncovering the cytochrome P450-catalyzed methylenedioxy bridge formation in streptovaricins biosynthesis.
Nat Commun, 11, 2020
|
|
1IVG
 
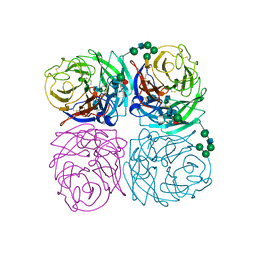 | | STRUCTURES OF AROMATIC INHIBITORS OF INFLUENZA VIRUS NEURAMINIDASE | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, INFLUENZA A SUBTYPE N2 NEURAMINIDASE, ... | | Authors: | Jedrzejas, M.J, Luo, M. | | Deposit date: | 1994-12-12 | | Release date: | 1995-03-31 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structures of aromatic inhibitors of influenza virus neuraminidase.
Biochemistry, 34, 1995
|
|
1RMF
 
 | |
1IVF
 
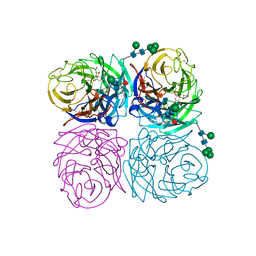 | | STRUCTURES OF AROMATIC INHIBITORS OF INFLUENZA VIRUS NEURAMINIDASE | | Descriptor: | 2-DEOXY-2,3-DEHYDRO-N-ACETYL-NEURAMINIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Jedrzejas, M.J, Luo, M. | | Deposit date: | 1994-12-12 | | Release date: | 1995-03-31 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structures of aromatic inhibitors of influenza virus neuraminidase.
Biochemistry, 34, 1995
|
|
1T7S
 
 | | Structural Genomics of Caenorhabditis elegans: Structure of BAG-1 protein | | Descriptor: | BAG-1 cochaperone | | Authors: | Symersky, J, Zhang, Y, Schormann, N, Li, S, Bunzel, R, Pruett, P, Luan, C.-H, Luo, M, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2004-05-10 | | Release date: | 2004-05-18 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural genomics of Caenorhabditis elegans: structure of the BAG domain.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
1VCJ
 
 | | Influenza B virus neuraminidase complexed with 1-(4-Carboxy-2-(3-pentylamino)phenyl)-5-aminomethyl-5-hydroxymethyl-pyrrolidin-2-one | | Descriptor: | 4-[(2R)-2-(AMINOMETHYL)-2-(HYDROXYMETHYL)-5-OXOPYRROLIDIN-1-YL]-3-[(1-ETHYLPROPYL)AMINO]BENZOIC ACID, NEURAMINIDASE | | Authors: | Lommer, B.S, Ali, S.M, Bajpai, S.N, Brouillette, W.J, Air, G.M, Luo, M. | | Deposit date: | 2004-03-09 | | Release date: | 2004-03-23 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | A benzoic acid inhibitor induces a novel conformational change in the active site of Influenza B virus neuraminidase.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
5U07
 
 | | CRISPR RNA-guided surveillance complex | | Descriptor: | CRISPR-associated protein, Cas5e family, Cse1 family, ... | | Authors: | Xiao, Y, Luo, M, Hayes, R.P, Kim, J, Ng, S, Ding, F, Liao, M, Ke, A. | | Deposit date: | 2016-11-23 | | Release date: | 2017-08-09 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structure Basis for Directional R-loop Formation and Substrate Handover Mechanisms in Type I CRISPR-Cas System.
Cell, 170, 2017
|
|
1AUA
 
 | | PHOSPHATIDYLINOSITOL TRANSFER PROTEIN SEC14P FROM SACCHAROMYCES CEREVISIAE | | Descriptor: | PHOSPHATIDYLINOSITOL TRANSFER PROTEIN SEC14P, octyl beta-D-glucopyranoside | | Authors: | Sha, B, Phillips, S.E, Bankaitis, V.A, Luo, M. | | Deposit date: | 1997-08-20 | | Release date: | 1997-12-24 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of the Saccharomyces cerevisiae phosphatidylinositol-transfer protein.
Nature, 391, 1998
|
|
6BJY
 
 | | VSV Nucleocapsid with Polyamide Bound | | Descriptor: | 4-{[4-(acetylamino)-1-methyl-1H-pyrrole-2-carbonyl]amino}-1-methyl-N-{4-[(1-methyl-1H-pyrrol-3-yl)amino]-4-oxobutyl}-1H-imidazole-2-carboxamide, Nucleoprotein, RNA (45-MER), ... | | Authors: | Gumpper, R.H, Luo, M. | | Deposit date: | 2017-11-07 | | Release date: | 2018-02-28 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.46 Å) | | Cite: | A Polyamide Inhibits Replication of Vesicular Stomatitis Virus by Targeting RNA in the Nucleocapsid.
J. Virol., 92, 2018
|
|
1AA7
 
 | |
1B9V
 
 | | NOVEL AROMATIC INHIBITORS OF INFLUENZA VIRUS NEURAMINIDASE MAKE SELECTIVE INTERACTIONS WITH CONSERVED RESIDUES AND WATER MOLECULES IN TEH ACTIVE SITE | | Descriptor: | 1-[4-CARBOXY-2-(3-PENTYLAMINO)PHENYL]-5,5'-DI(HYDROXYMETHYL)PYRROLIDIN-2-ONE, 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Finley, J.B, Atigadda, V.R, Duarte, F, Zahao, J.J, Brouillette, W.J, Air, G.M, Luo, M. | | Deposit date: | 1999-02-15 | | Release date: | 1999-02-27 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Novel aromatic inhibitors of influenza virus neuraminidase make selective interactions with conserved residues and water molecules in the active site.
J.Mol.Biol., 293, 1999
|
|
5U0A
 
 | | CRISPR RNA-guided surveillance complex | | Descriptor: | CRISPR-associated protein, Cas5e family, Cse1 family, ... | | Authors: | Xiao, Y, Luo, M, Hayes, R.P, Kim, J, Ng, S, Ding, F, Liao, M, Ke, A. | | Deposit date: | 2016-11-23 | | Release date: | 2017-08-09 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structure Basis for Directional R-loop Formation and Substrate Handover Mechanisms in Type I CRISPR-Cas System.
Cell, 170, 2017
|
|
6BB5
 
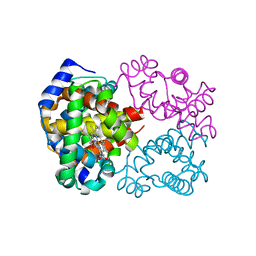 | | Human Oxy-Hemoglobin | | Descriptor: | Hemoglobin subunit alpha, Hemoglobin subunit beta, OXYGEN MOLECULE, ... | | Authors: | Gumpper, R.H, Terrell, J.R, Luo, M. | | Deposit date: | 2017-10-16 | | Release date: | 2018-01-10 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Hemoglobin crystals immersed in liquid oxygen reveal diffusion channels.
Biochem. Biophys. Res. Commun., 495, 2018
|
|
1B9S
 
 | | NOVEL AROMATIC INHIBITORS OF INFLUENZA VIRUS NEURAMINIDASE MAKE SELECTIVE INTERACTIONS WITH CONSERVED RESIDUES AND WATER MOLECULES IN THE ACTIVE SITE | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-(N-ACETYLAMINO)-3-[N-(2-ETHYLBUTANOYLAMINO)]BENZOIC ACID, CALCIUM ION, ... | | Authors: | Finley, J.B, Atigadda, V.R, Duarte, F, Zhao, J.J, Brouillette, W.J, Air, G.M, Luo, M. | | Deposit date: | 1999-02-15 | | Release date: | 1999-02-26 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Novel aromatic inhibitors of influenza virus neuraminidase make selective interactions with conserved residues and water molecules in the active site.
J.Mol.Biol., 293, 1999
|
|
1B9T
 
 | | NOVEL AROMATIC INHIBITORS OF INFLUENZA VIRUS NEURAMINIDASE MAKE SELECTIVE INTERACTIONS WITH CONSERVED RESIDUES AND WATER MOLECULES IN THE ACTIVE SITE | | Descriptor: | 1-(4-CARBOXY-2-GUANIDINOPENTYL)-5,5'-DI(HYDROXYMETHYL)PYRROLIDIN-2-ONE, 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Finley, J.B, Atigadda, V.R, Duarte, F, Zhao, J.J, Brouillette, W.J, Air, G.M, Luo, M. | | Deposit date: | 1999-02-15 | | Release date: | 1999-02-27 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Novel aromatic inhibitors of influenza virus neuraminidase make selective interactions with conserved residues and water molecules in the active site.
J.Mol.Biol., 293, 1999
|
|
4Q71
 
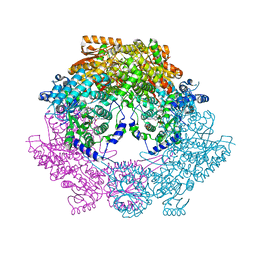 | | Crystal Structure of Bradyrhizobium japonicum Proline Utilization A (PutA) Mutant D779W | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, Proline dehydrogenase, ... | | Authors: | Tanner, J.J, Luo, M, Pemberton, T.A. | | Deposit date: | 2014-04-23 | | Release date: | 2014-08-06 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Kinetic and Structural Characterization of Tunnel-Perturbing Mutants in Bradyrhizobium japonicum Proline Utilization A.
Biochemistry, 53, 2014
|
|
4Q72
 
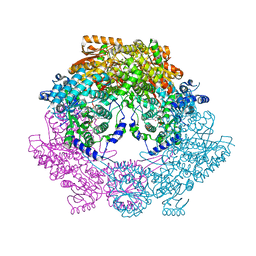 | | Crystal Structure of Bradyrhizobium japonicum Proline Utilization A (PutA) Mutant D779Y | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, Proline dehydrogenase, ... | | Authors: | Tanner, J.J, Pemberton, T.A, Luo, M. | | Deposit date: | 2014-04-23 | | Release date: | 2014-08-06 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Kinetic and Structural Characterization of Tunnel-Perturbing Mutants in Bradyrhizobium japonicum Proline Utilization A.
Biochemistry, 53, 2014
|
|
4Q73
 
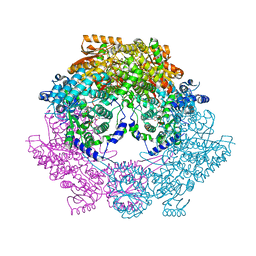 | | Crystal Structure of Bradyrhizobium japonicum Proline Utilization A (PutA) Mutant D778Y | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, Proline dehydrogenase, ... | | Authors: | Tanner, J.J, Luo, M, Pemberton, T.A. | | Deposit date: | 2014-04-23 | | Release date: | 2014-08-06 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Kinetic and Structural Characterization of Tunnel-Perturbing Mutants in Bradyrhizobium japonicum Proline Utilization A.
Biochemistry, 53, 2014
|
|
3PIN
 
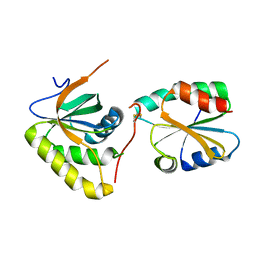 | | Crystal structure of Mxr1 from Saccharomyces cerevisiae in complex with Trx2 | | Descriptor: | Peptide methionine sulfoxide reductase, Thioredoxin-2 | | Authors: | Ma, X.X, Guo, P.C, Shi, W.W, Luo, M, Tan, X.F, Chen, Y, Zhou, C.Z. | | Deposit date: | 2010-11-07 | | Release date: | 2011-02-23 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural plasticity of the thioredoxin recognition site of yeast methionine S-sulfoxide reductase Mxr1
J.Biol.Chem., 286, 2011
|
|
3PIM
 
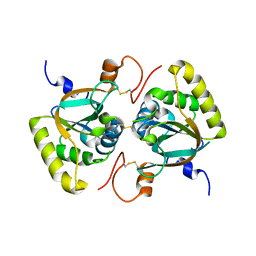 | | Crystal structure of Mxr1 from Saccharomyces cerevisiae in unusual oxidized form | | Descriptor: | Peptide methionine sulfoxide reductase | | Authors: | Ma, X.X, Guo, P.C, Shi, W.W, Luo, M, Tan, X.F, Chen, Y, Zhou, C.Z. | | Deposit date: | 2010-11-07 | | Release date: | 2011-02-23 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural plasticity of the thioredoxin recognition site of yeast methionine S-sulfoxide reductase Mxr1
J.Biol.Chem., 286, 2011
|
|
