9E9H
 
 | | Crystal structure of human KRAS G12C covalently bound to DEL triazine compound 5 | | Descriptor: | (3S)-N,5-dimethyl-3-({4-[3-(morpholin-4-yl)phenyl]-6-(2-propanoyl-2,6-diazaspiro[3.4]octan-6-yl)-1,3,5-triazin-2-yl}amino)hexanamide, CALCIUM ION, GTPase KRas, ... | | Authors: | Mohr, C. | | Deposit date: | 2024-11-08 | | Release date: | 2025-02-26 | | Last modified: | 2025-03-05 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Identification of Structurally Novel KRAS G12C Inhibitors through Covalent DNA-Encoded Library Screening.
J.Med.Chem., 68, 2025
|
|
9E9I
 
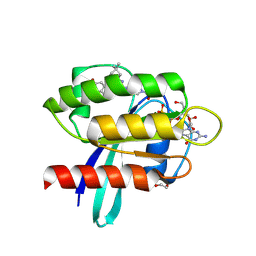 | | Crystal Structure of human KRAS G12C covalently bound to nopinone-derived naphthol compound 21 | | Descriptor: | 1,2-ETHANEDIOL, 1-{6-[(4P,6R,8R)-3-fluoro-4-(3-hydroxynaphthalen-1-yl)-7,7-dimethyl-5,6,7,8-tetrahydro-6,8-methanoquinolin-2-yl]-2,6-diazaspiro[3.4]octan-2-yl}propan-1-one, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Mohr, C. | | Deposit date: | 2024-11-08 | | Release date: | 2025-02-26 | | Last modified: | 2025-03-05 | | Method: | X-RAY DIFFRACTION (1.18 Å) | | Cite: | Identification of Structurally Novel KRAS G12C Inhibitors through Covalent DNA-Encoded Library Screening.
J.Med.Chem., 68, 2025
|
|
7TDX
 
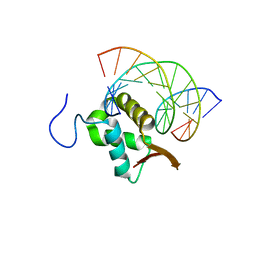 | | Structure of FOXP3-DNA complex | | Descriptor: | DNA (5'-D(*AP*AP*AP*TP*TP*TP*GP*TP*TP*TP*AP*CP*TP*C)-3'), DNA (5'-D(P*GP*AP*GP*TP*AP*AP*AP*CP*AP*AP*AP*TP*TP*T)-3'), Forkhead box P3 | | Authors: | Leng, F, Hur, S. | | Deposit date: | 2022-01-03 | | Release date: | 2022-08-10 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | The transcription factor FoxP3 can fold into two dimerization states with divergent implications for regulatory T cell function and immune homeostasis.
Immunity, 55, 2022
|
|
7TDW
 
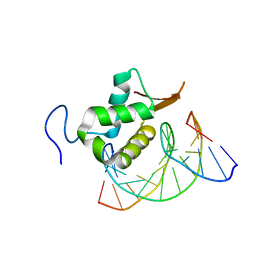 | | Structure of FOXP3-DNA complex | | Descriptor: | DNA (5'-D(*AP*AP*AP*TP*TP*TP*GP*TP*TP*TP*AP*CP*TP*C)-3'), DNA (5'-D(P*GP*AP*GP*TP*AP*AP*AP*CP*AP*AP*AP*TP*TP*T)-3'), Forkhead box P3 | | Authors: | Leng, F, Hur, S. | | Deposit date: | 2022-01-03 | | Release date: | 2022-08-10 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (4 Å) | | Cite: | The transcription factor FoxP3 can fold into two dimerization states with divergent implications for regulatory T cell function and immune homeostasis.
Immunity, 55, 2022
|
|
6VOY
 
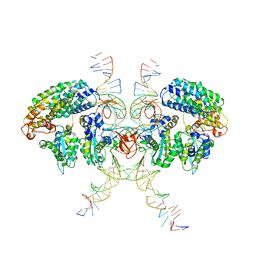 | | Cryo-EM structure of HTLV-1 instasome | | Descriptor: | DNA (25-MER), DNA (5'-D(P*AP*CP*AP*CP*AP*CP*TP*TP*GP*AP*CP*TP*AP*GP*GP*GP*TP*G)-3'), DNA-binding protein 7d, ... | | Authors: | Bhatt, V, Shi, K, Sundborger, A, Aihara, H. | | Deposit date: | 2020-02-01 | | Release date: | 2020-07-01 | | Last modified: | 2025-05-28 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structural basis of host protein hijacking in human T-cell leukemia virus integration.
Nat Commun, 11, 2020
|
|
5B76
 
 | |
5B77
 
 | |
6DCH
 
 | | Structure of isonitrile biosynthesis enzyme ScoE | | Descriptor: | ACETATE ION, CHLORIDE ION, CHOLINE ION, ... | | Authors: | Born, D.A, Drennan, C.L. | | Deposit date: | 2018-05-07 | | Release date: | 2018-06-27 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Isonitrile Formation by a Non-Heme Iron(II)-Dependent Oxidase/Decarboxylase.
Angew. Chem. Int. Ed. Engl., 57, 2018
|
|
8D4X
 
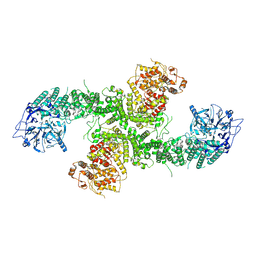 | | Structure of the human UBR5 HECT-type E3 ubiquitin ligase in a dimeric form | | Descriptor: | E3 ubiquitin-protein ligase UBR5, ZINC ION | | Authors: | Wang, F, He, Q, Lin, G, Li, H. | | Deposit date: | 2022-06-02 | | Release date: | 2023-04-19 | | Last modified: | 2025-05-28 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structure of the human UBR5 E3 ubiquitin ligase.
Structure, 31, 2023
|
|
7TV2
 
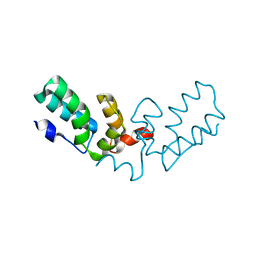 | | X-ray crystal structure of HIV-2 CA protein CTD | | Descriptor: | Capsid protein p24 | | Authors: | Shi, K, Aihara, H. | | Deposit date: | 2022-02-03 | | Release date: | 2023-05-17 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | HIV-2 Immature Particle Morphology Provides Insights into Gag Lattice Stability and Virus Maturation.
J.Mol.Biol., 435, 2023
|
|
6WAU
 
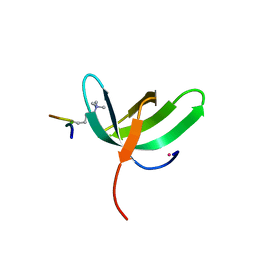 | | Complex structure of PHF19 | | Descriptor: | Histone H3.1t peptide, PHD finger protein 19, UNKNOWN ATOM OR ION | | Authors: | Dong, C, Bountra, C, Edwards, A.M, Arrowsmith, C.H, Min, J.R, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-03-26 | | Release date: | 2020-08-26 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural basis for histone variant H3tK27me3 recognition by PHF1 and PHF19.
Elife, 9, 2020
|
|
6WAV
 
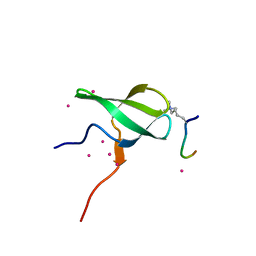 | | Crystal structure of PHF1 in complex with H3K36me3 substitution | | Descriptor: | Histone H3.1, PHD finger protein 1, SULFATE ION, ... | | Authors: | Dong, C, Bountra, C, Edwards, A.M, Arrowsmith, C.H, Min, J.R, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-03-26 | | Release date: | 2020-08-26 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural basis for histone variant H3tK27me3 recognition by PHF1 and PHF19.
Elife, 9, 2020
|
|
6WAT
 
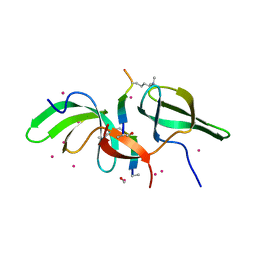 | | complex structure of PHF1 | | Descriptor: | Histone H3.1t peptide, PHD finger protein 1, UNKNOWN ATOM OR ION | | Authors: | Dong, C, Bountra, C, Edwards, A.M, Arrowsmith, C.H, Min, J.R, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-03-26 | | Release date: | 2020-08-26 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis for histone variant H3tK27me3 recognition by PHF1 and PHF19.
Elife, 9, 2020
|
|
5BW0
 
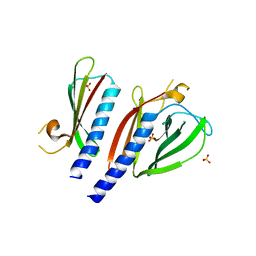 | | The crystal structure of minor pseudopilin binary complex of XcpV and XcpW from the Type 2 secretion system of Pseudomonas aeruginosa | | Descriptor: | SULFATE ION, Type II secretion system protein I, Type II secretion system protein J | | Authors: | Zhang, Y, Faucher, F, Poole, K, Jia, Z. | | Deposit date: | 2015-06-05 | | Release date: | 2016-07-20 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure-guided disruption of the pseudopilus tip complex inhibits the Type II secretion in Pseudomonas aeruginosa.
PLoS Pathog., 14, 2018
|
|
5B78
 
 | |
6KEV
 
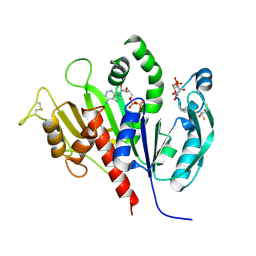 | | Reduced phosphoribulokinase from Synechococcus elongatus PCC 7942 complexed with adenosine diphosphate and glucose 6-phosphate | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, 6-O-phosphono-alpha-D-glucopyranose, ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Yu, A, Xie, Y, Li, M. | | Deposit date: | 2019-07-05 | | Release date: | 2020-05-13 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.506139 Å) | | Cite: | Photosynthetic Phosphoribulokinase Structures: Enzymatic Mechanisms and the Redox Regulation of the Calvin-Benson-Bassham Cycle.
Plant Cell, 32, 2020
|
|
5B79
 
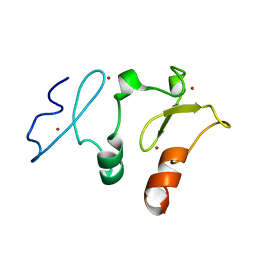 | | Crystal structure of DPF2 double PHD finger | | Descriptor: | ZINC ION, Zinc finger protein ubi-d4 | | Authors: | Li, H, Xiong, X. | | Deposit date: | 2016-06-05 | | Release date: | 2016-10-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.601 Å) | | Cite: | Selective recognition of histone crotonylation by double PHD fingers of MOZ and DPF2
Nat.Chem.Biol., 12, 2016
|
|
3N2Y
 
 | |
6KEW
 
 | |
6KEZ
 
 | | Crystal structure of GAPDH/CP12/PRK complex from Arabidopsis thaliana | | Descriptor: | Calvin cycle protein CP12-2, Glyceraldehyde-3-phosphate dehydrogenase GAPA1, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Yu, A, Xie, Y, Li, M. | | Deposit date: | 2019-07-05 | | Release date: | 2020-05-13 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Photosynthetic Phosphoribulokinase Structures: Enzymatic Mechanisms and the Redox Regulation of the Calvin-Benson-Bassham Cycle.
Plant Cell, 32, 2020
|
|
6KEX
 
 | |
6ID0
 
 | | Cryo-EM structure of a human intron lariat spliceosome prior to Prp43 loaded (ILS1 complex) at 2.9 angstrom resolution | | Descriptor: | 116 kDa U5 small nuclear ribonucleoprotein component, CWF19-like protein 2, Cell division cycle 5-like protein, ... | | Authors: | Zhang, X, Zhan, X, Yan, C, Shi, Y. | | Deposit date: | 2018-09-07 | | Release date: | 2019-03-13 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structures of the human spliceosomes before and after release of the ligated exon.
Cell Res., 29, 2019
|
|
8X6Y
 
 | | Crystal structure of EfCDA | | Descriptor: | CACODYLATE ION, EfCDA, ZINC ION | | Authors: | Jiang, L, Huang, Y. | | Deposit date: | 2023-11-22 | | Release date: | 2024-11-27 | | Last modified: | 2025-06-11 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Deciphering the role of Enterococcus faecium cytidine deaminase in gemcitabine resistance of gallbladder cancer.
J.Biol.Chem., 300, 2024
|
|
8X6W
 
 | | Crystal structure of EfCDA | | Descriptor: | EfCDA, ZINC ION | | Authors: | Jiang, L, Huang, Y. | | Deposit date: | 2023-11-22 | | Release date: | 2024-11-27 | | Last modified: | 2025-06-11 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Deciphering the role of Enterococcus faecium cytidine deaminase in gemcitabine resistance of gallbladder cancer.
J.Biol.Chem., 300, 2024
|
|
8X6U
 
 | | Crystal structure of EfCDA in complex with dFdU | | Descriptor: | 2',2'-Difluorodeoxyuridine, EfCDA, ZINC ION | | Authors: | Jiang, L, Huang, Y. | | Deposit date: | 2023-11-22 | | Release date: | 2024-11-27 | | Last modified: | 2025-06-11 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Deciphering the role of Enterococcus faecium cytidine deaminase in gemcitabine resistance of gallbladder cancer.
J.Biol.Chem., 300, 2024
|
|
