7EGS
 
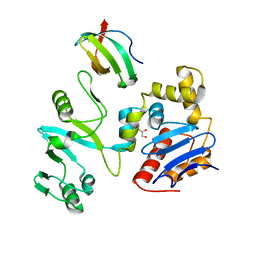 | | The crystal structure of lobe domain of E. coli RNA polymerase complexed with the C-terminal domain of UvrD | | Descriptor: | DNA helicase II, DNA-directed RNA polymerase subunit beta, GLYCEROL | | Authors: | Zheng, F, Shen, L, Li, L, Zhang, Y. | | Deposit date: | 2021-03-26 | | Release date: | 2022-04-06 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crucial role and mechanism of transcription-coupled DNA repair in bacteria.
Nature, 604, 2022
|
|
7EGT
 
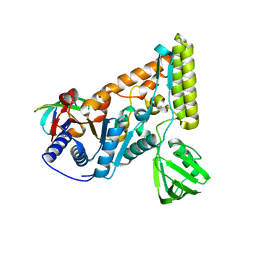 | | The crystal structure of the C-terminal domain of T. thermophilus UvrD complexed with the N-terminal domain of UvrB | | Descriptor: | DNA helicase UvrD, UvrABC system protein B | | Authors: | Zheng, F, Shen, L, Li, L, Zhang, Y. | | Deposit date: | 2021-03-26 | | Release date: | 2022-04-06 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.581 Å) | | Cite: | Crucial role and mechanism of transcription-coupled DNA repair in bacteria.
Nature, 604, 2022
|
|
5DP4
 
 | | Crystal Structure of EV71 3C Proteinase in complex with compound 3 | | Descriptor: | 3C proteinase, ethyl (2Z,4S)-4-{[(2S)-2-methyl-3-phenylpropanoyl]amino}-5-[(3S)-2-oxopyrrolidin-3-yl]pent-2-enoate | | Authors: | Wu, C, Zhang, L, Li, P, Cai, Q, Peng, X, Li, N, Cai, Y, Li, J, Lin, T. | | Deposit date: | 2015-09-12 | | Release date: | 2016-03-30 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Fragment-wise design of inhibitors to 3C proteinase from enterovirus 71
Biochim.Biophys.Acta, 1860, 2016
|
|
5DJO
 
 | | Crystal structure of the CC1-FHA tandem of Kinesin-3 KIF13A | | Descriptor: | ACETIC ACID, DI(HYDROXYETHYL)ETHER, FORMIC ACID, ... | | Authors: | Ren, J.Q, Li, W, Huo, L, Feng, W. | | Deposit date: | 2015-09-02 | | Release date: | 2015-12-30 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Structural Correlation of the Neck Coil with the Coiled-coil (CC1)-Forkhead-associated (FHA) Tandem for Active Kinesin-3 KIF13A
J.Biol.Chem., 291, 2016
|
|
1U15
 
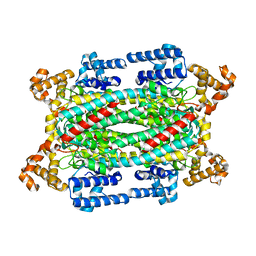 | | Crystal structure of a duck-delta-crystallin-1 double loop mutant (DLM) | | Descriptor: | Delta crystallin I | | Authors: | Tsai, M, Sampaleanu, L.M, Greene, C, Creagh, L, Haynes, C, Howell, P.L. | | Deposit date: | 2004-07-14 | | Release date: | 2004-10-05 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A duck delta1 crystallin double loop mutant provides insight into residues important for argininosuccinate lyase activity.
Biochemistry, 43, 2004
|
|
1XY8
 
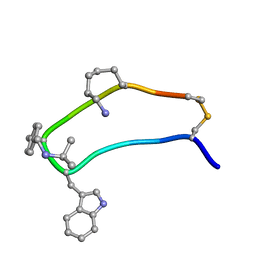 | | NMR strcutre of sst1-selective somatostatin (SRIF) analog 1 | | Descriptor: | SST1-selective somatosatin analog | | Authors: | Grace, C.R.R, Durrer, L, Koerber, S.C, Erchegyi, J, Reubi, J.C, Rivier, J.E, Riek, R. | | Deposit date: | 2004-11-09 | | Release date: | 2005-02-15 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | Somatostatin receptor 1 selective analogues: 4. Three-dimensional consensus structure by NMR
J.Med.Chem., 48, 2005
|
|
8ORS
 
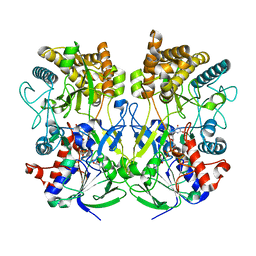 | | Knockout of GMC-oxidoreductase genes reveals that functional redundancy preserves mimivirus essential functions | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Putative GMC-type oxidoreductase | | Authors: | Alempic, J.M, Bisio, H, Villalta, A, Santini, S, Lartigue, A, Schmitt, A, Bugnot, C, Notaro, A, Belmudes, L, Adrait, A, Poirot, O, Ptchelkine, D, De Castro, C, Coute, Y, Abergel, C. | | Deposit date: | 2023-04-17 | | Release date: | 2024-04-17 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Functional redundancy revealed by the deletion of the mimivirus GMC-oxidoreductase genes.
Microlife, 5, 2024
|
|
1XYU
 
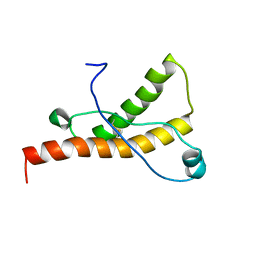 | | Solution structure of the sheep prion protein with polymorphism H168 | | Descriptor: | Major prion protein | | Authors: | Calzolai, L, Lysek, D.A, Guntert, P, Wuthrich, K. | | Deposit date: | 2004-11-11 | | Release date: | 2005-01-04 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | Prion protein NMR structures of cats, dogs, pigs, and sheep
Proc.Natl.Acad.Sci.USA, 102, 2005
|
|
1U1Y
 
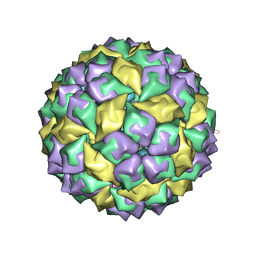 | | Crystal structure of a complex between WT bacteriophage MS2 coat protein and an F5 aptamer RNA stemloop with 2aminopurine substituted at the-10 position | | Descriptor: | 5'-R(*CP*CP*GP*GP*(2PR)P*GP*GP*AP*UP*CP*AP*CP*CP*AP*CP*GP*G)-3', Coat protein | | Authors: | Horn, W.T, Convery, M.A, Stonehouse, N.J, Adams, C.J, Liljas, L, Phillips, S.E, Stockley, P.G. | | Deposit date: | 2004-07-16 | | Release date: | 2004-12-07 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | The crystal structure of a high affinity RNA stem-loop complexed with the bacteriophage MS2 capsid: further challenges in the modeling of ligand-RNA interactions.
Rna, 10, 2004
|
|
8P1U
 
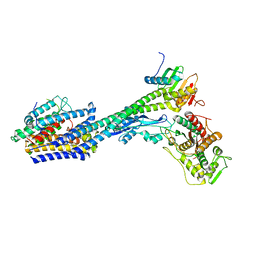 | | Structure of divisome complex FtsWIQLB | | Descriptor: | Cell division protein FtsB, Cell division protein FtsL, Cell division protein FtsQ, ... | | Authors: | Yang, L, Chang, S, Tang, D, Dong, H. | | Deposit date: | 2023-05-12 | | Release date: | 2024-05-22 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural insights into the activation of the divisome complex FtsWIQLB.
Cell Discov, 10, 2024
|
|
8OV9
 
 | |
1W0X
 
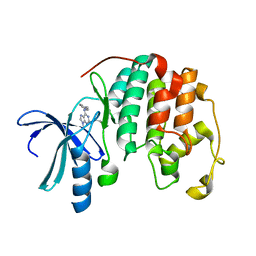 | |
8P2O
 
 | |
8P2P
 
 | |
1TUD
 
 | |
8P1K
 
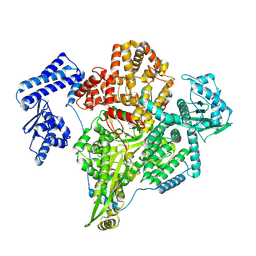 | |
8P1M
 
 | |
8OV8
 
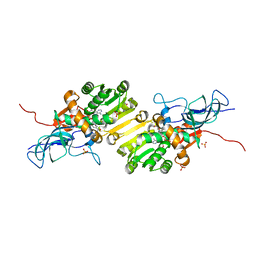 | | Crystal structure of Ene-reductase 1 from black poplar mushroom in complex to NADP | | Descriptor: | Ene-reductase 1, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, SODIUM ION, ... | | Authors: | Korf, L, Essen, L.-O, Karrer, D, Ruehl, M. | | Deposit date: | 2023-04-25 | | Release date: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Shifting the substrate scope of an ene/yne-reductase by loop engineering
To Be Published
|
|
1W5V
 
 | | HIV-1 protease in complex with fluoro substituted diol-based C2- symmetric inhibitor | | Descriptor: | HIV-1 PROTEASE, N,N-[2,5-O-DI-3-FLUORO-BENZYL-GLUCARYL]-DI-[1-AMINO-INDAN-2-OL] | | Authors: | Lindberg, J, Pyring, D, Loewgren, S, Rosenquist, A, Zuccarello, G, Kvarnstroem, I, Zhang, H, Vrang, L, Claesson, B, Hallberg, A, Samuelsson, B, Unge, T. | | Deposit date: | 2004-08-10 | | Release date: | 2004-12-01 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Symmetric Fluoro-Substituted Diol-Based HIV Protease Inhibitors. Ortho-Fluorinated and Meta-Fluorinated P1/P1'-Benzyloxy Side Groups Significantly Improve the Antiviral Activity and Preserve Binding Efficacy
Eur.J.Biochem., 271, 2004
|
|
6CB1
 
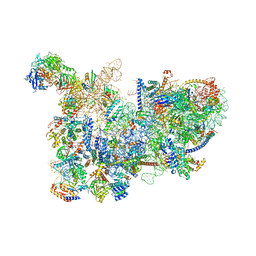 | | Yeast nucleolar pre-60S ribosomal subunit (state 3) | | Descriptor: | 35S pre-ribosomal RNA miscRNA, 5.8S rRNA, 60S ribosomal protein L13-A, ... | | Authors: | Sanghai, Z.A, Miller, L, Barandun, J, Hunziker, M, Chaker-Margot, M, Klinge, S. | | Deposit date: | 2018-02-01 | | Release date: | 2018-03-14 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (4.6 Å) | | Cite: | Modular assembly of the nucleolar pre-60S ribosomal subunit.
Nature, 556, 2018
|
|
8P2N
 
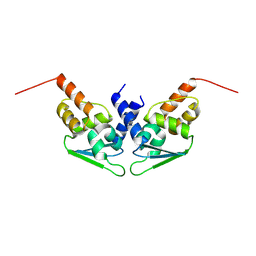 | |
5DVX
 
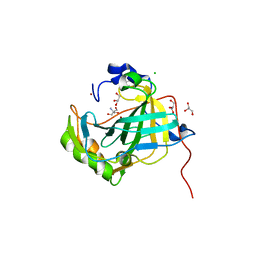 | | Crystal structure of the catalytic-domain of human carbonic anhydrase IX at 1.6 angstrom resolution | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, Carbonic anhydrase 9, ... | | Authors: | Mahon, B.P, Socorro, L, Driscoll, J.M, McKenna, R. | | Deposit date: | 2015-09-21 | | Release date: | 2016-08-03 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.598 Å) | | Cite: | The Structure of Carbonic Anhydrase IX Is Adapted for Low-pH Catalysis.
Biochemistry, 55, 2016
|
|
1TT4
 
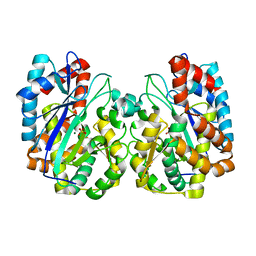 | | Structure of NP459575, a predicted glutathione synthase from Salmonella typhimurium | | Descriptor: | MAGNESIUM ION, SULFATE ION, putative cytoplasmic protein | | Authors: | Miller, D.J, Shuvalova, L, Anderson, W.F, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2004-06-21 | | Release date: | 2004-08-17 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.801 Å) | | Cite: | Structure of NP459575, a predicted glutathione synthase from Salmonella typhimurium
To be Published
|
|
1WA9
 
 | | Crystal Structure of the PAS repeat region of the Drosophila clock protein PERIOD | | Descriptor: | PERIOD CIRCADIAN PROTEIN | | Authors: | Yildiz, O, Doi, M, Yujnovsky, I, Cardone, L, Berndt, A, Hennig, S, Schulze, S, Urbanke, C, Sassone-Corsi, P, Wolf, E. | | Deposit date: | 2004-10-25 | | Release date: | 2005-01-12 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Crystal Structure and Interactions of the Pas Repeat Region of the Drosophila Clock Protein Period
Mol.Cell, 17, 2005
|
|
5DXY
 
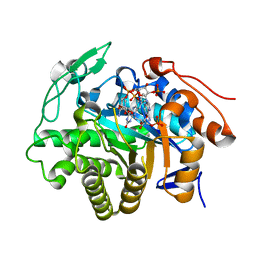 | | Crystal structure of Dbr2 | | Descriptor: | Artemisinic aldehyde Delta(11(13)) reductase, FLAVIN MONONUCLEOTIDE, [[(2R,3S,4R,5R)-5-(5-aminocarbonyl-3,4-dihydro-2H-pyridin-1-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methoxy-oxidanyl-phosphoryl] [(2R,3R,4R,5R)-5-(6-aminopurin-9-yl)-3-oxidanyl-4-phosphonooxy-oxolan-2-yl]methyl hydrogen phosphate | | Authors: | Li, L, Chen, T.T, Xu, Y.C. | | Deposit date: | 2015-09-24 | | Release date: | 2016-10-05 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | The Crystal structure of Dbr2
to be published
|
|
