6DWF
 
 | |
4RLD
 
 | |
6DWU
 
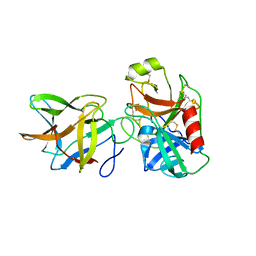 | | Crystal structure of complex of BBKI and Bovine Trypsin | | Descriptor: | Cationic trypsin, Kunitz-type inihibitor | | Authors: | Li, M, Wlodawer, A, Gustchina, A. | | Deposit date: | 2018-06-28 | | Release date: | 2019-01-30 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.96 Å) | | Cite: | Crystal structures of the complex of a kallikrein inhibitor from Bauhinia bauhinioides with trypsin and modeling of kallikrein complexes.
Acta Crystallogr D Struct Biol, 75, 2019
|
|
6DF3
 
 | |
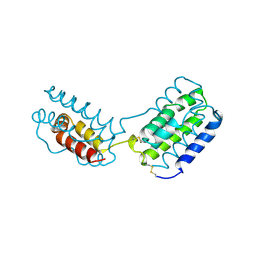
1VLK
 
 | |
7R6A
 
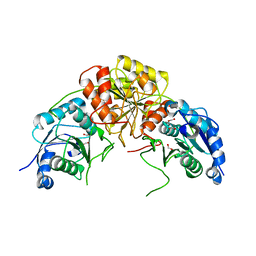 | |
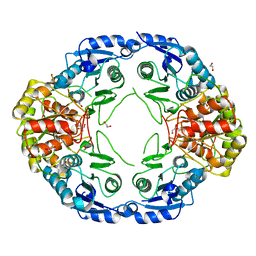
7R69
 
 | |
7R6B
 
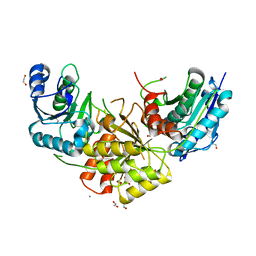 | | Crystal structure of mutant R43D/L124D/R125A/C273S of L-Asparaginase I from Yersinia pestis | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, L-asparaginase I | | Authors: | Strzelczyk, P, Wlodawer, A, Lubkowski, J. | | Deposit date: | 2021-06-22 | | Release date: | 2022-07-06 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | The dimeric form of bacterial l-asparaginase YpAI is fully active.
Febs J., 290, 2023
|
|
3TLH
 
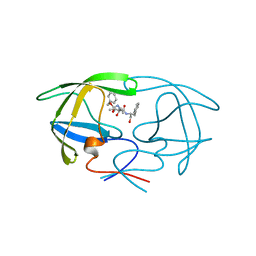 | | STRUCTURAL STUDIES OF HIV AND FIV PROTEASES COMPLEXED WITHAN EFFICIENT INHIBITOR OF FIV PR | | Descriptor: | PROTEIN (PROTEASE), benzyl [(1S,4S,7S,8R,9R,10S,13S,16S)-7,10-dibenzyl-8,9-dihydroxy-1,16-dimethyl-4,13-bis(1-methylethyl)-2,5,12,15,18-pentaoxo-20-phenyl-19-oxa-3,6,11,14,17-pentaazaicos-1-yl]carbamate | | Authors: | Li, M, Lee, T, Morris, G, Laco, G, Wong, C, Olson, A, Elder, J, Wlodawer, A, Gustchina, A. | | Deposit date: | 1998-12-03 | | Release date: | 1998-12-09 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural studies of FIV and HIV-1 proteases complexed with an efficient inhibitor of FIV protease
Proteins, 38, 2000
|
|
9ARQ
 
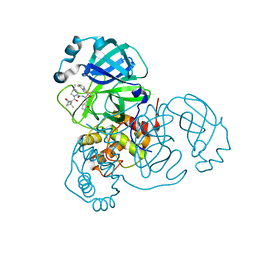 | | Crystal structure of SARS-CoV-2 main protease (authentic protein) in complex with an inhibitor TKB-245 | | Descriptor: | (1R,2S,5S)-N-{(1S,2S)-1-(4-fluoro-1,3-benzothiazol-2-yl)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase nsp5 | | Authors: | Bulut, H, Hattori, S, Hayashi, H, Hasegawa, K, Li, M, Wlodawer, A, Tamamura, H, Mitsuya, H. | | Deposit date: | 2024-02-23 | | Release date: | 2024-04-24 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural and virologic mechanism of the emergence of resistance to M pro inhibitors in SARS-CoV-2.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
9ARS
 
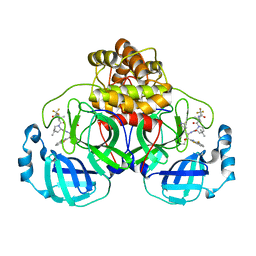 | | Crystal structure of SARS-CoV-2 main protease E166V mutant in complex with an inhibitor TKB-245 | | Descriptor: | (1R,2S,5S)-N-{(1S,2S)-1-(4-fluoro-1,3-benzothiazol-2-yl)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase nsp5 | | Authors: | Bulut, H, Hattori, S, Hayashi, H, Hasegawa, K, Li, M, Wlodawer, A, Misumi, S, Tamamura, H, Mitsuya, H. | | Deposit date: | 2024-02-23 | | Release date: | 2024-04-24 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural and virologic mechanism of the emergence of resistance to M pro inhibitors in SARS-CoV-2.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
9ART
 
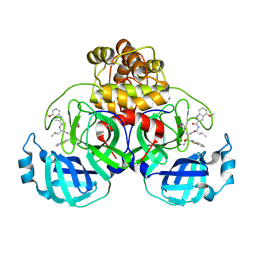 | | Crystal structure of SARS-CoV-2 main protease A191T mutant in complex with an inhibitor 5h | | Descriptor: | 3C-like proteinase nsp5, N-[(2S)-1-({(1S,2S)-1-(1,3-benzothiazol-2-yl)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}amino)-4-methyl-1-oxopentan-2-yl]-4-methoxy-1H-indole-2-carboxamide | | Authors: | Bulut, H, Hattori, S, Hayashi, H, Hasegawa, K, Li, M, Wlodawer, A, Tamamura, H, Mitsuya, H. | | Deposit date: | 2024-02-23 | | Release date: | 2024-04-24 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Structural and virologic mechanism of the emergence of resistance to M pro inhibitors in SARS-CoV-2.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
9AVQ
 
 | | Crystal structure of SARS-CoV-2 main protease A191T mutant in complex with an inhibitor Nirmatrelvir | | Descriptor: | (1R,2S,5S)-N-{(1E,2S)-1-imino-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase nsp5, DI(HYDROXYETHYL)ETHER | | Authors: | Bulut, H, Hattori, S, Hayashi, H, Hasegawa, K, Li, M, Wlodawer, A, Tamamura, H, Mitsuya, H. | | Deposit date: | 2024-03-04 | | Release date: | 2024-04-24 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.58 Å) | | Cite: | Structural and virologic mechanism of the emergence of resistance to M pro inhibitors in SARS-CoV-2.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
6DWH
 
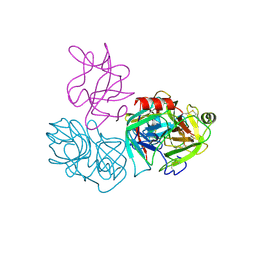 | | Crystal structure of complex of BBKI and Bovine Trypsin | | Descriptor: | CHLORIDE ION, Cationic trypsin, Kunitz-type inihibitor, ... | | Authors: | Li, M, Wlodawer, A. | | Deposit date: | 2018-06-26 | | Release date: | 2019-01-30 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structures of the complex of a kallikrein inhibitor from Bauhinia bauhinioides with trypsin and modeling of kallikrein complexes.
Acta Crystallogr D Struct Biol, 75, 2019
|
|
2ITG
 
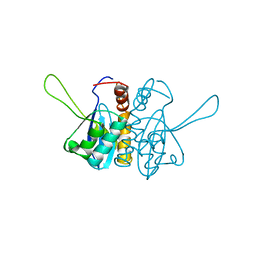 | | CATALYTIC DOMAIN OF HIV-1 INTEGRASE: ORDERED ACTIVE SITE IN THE F185H CONSTRUCT | | Descriptor: | HUMAN IMMUNODEFICIENCY VIRUS-1 INTEGRASE | | Authors: | Bujacz, G, Alexandratos, J, Wlodawer, A, Zhou-Liu, Q, Clement-Mella, C. | | Deposit date: | 1996-09-13 | | Release date: | 1997-03-12 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The catalytic domain of human immunodeficiency virus integrase: ordered active site in the F185H mutant.
FEBS Lett., 398, 1996
|
|
8ECE
 
 | | E. coli L-asparaginase II mutant (V27T) in complex with L-Glu | | Descriptor: | 1,2-ETHANEDIOL, GLUTAMIC ACID, L-asparaginase 2 | | Authors: | Strzelczyk, P, Wlodawer, A, Lubkowski, J. | | Deposit date: | 2022-09-01 | | Release date: | 2022-11-16 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | The E. coli L-asparaginase V27T mutant: structural and functional characterization and comparison with theoretical predictions.
Febs Lett., 596, 2022
|
|
8ECD
 
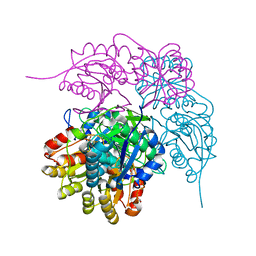 | | E. coli L-asparaginase II mutant (V27T) in complex with L-Asp | | Descriptor: | 1,2-ETHANEDIOL, ASPARTIC ACID, CITRIC ACID, ... | | Authors: | Strzelczyk, P, Wlodawer, A, Lubkowski, J. | | Deposit date: | 2022-09-01 | | Release date: | 2022-11-16 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | The E. coli L-asparaginase V27T mutant: structural and functional characterization and comparison with theoretical predictions.
Febs Lett., 596, 2022
|
|
3ECA
 
 | | CRYSTAL STRUCTURE OF ESCHERICHIA COLI L-ASPARAGINASE, AN ENZYME USED IN CANCER THERAPY (ELSPAR) | | Descriptor: | ASPARTIC ACID, L-asparaginase 2 | | Authors: | Swain, A.L, Jaskolski, M, Housset, D, Rao, J.K.M, Wlodawer, A. | | Deposit date: | 1993-07-02 | | Release date: | 1993-10-31 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of Escherichia coli L-asparaginase, an enzyme used in cancer therapy.
Proc.Natl.Acad.Sci.USA, 90, 1993
|
|
5C9B
 
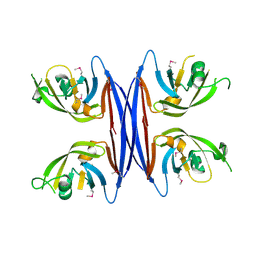 | | Crystal structure of a retropepsin-like aspartic protease from Rickettsia conorii | | Descriptor: | ApRick protease | | Authors: | Li, M, Gustchina, A, Cruz, R, Simoes, M, Curto, P, Martinez, J, Faro, C, Simoes, I, Wlodawer, A. | | Deposit date: | 2015-06-26 | | Release date: | 2015-10-14 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of RC1339/APRc from Rickettsia conorii, a retropepsin-like aspartic protease.
Acta Crystallogr. D Biol. Crystallogr., 71, 2015
|
|
4YDX
 
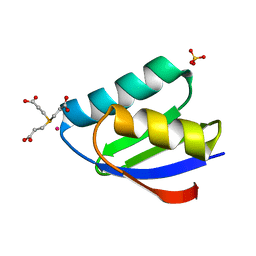 | | Crystal structure of cisplatin bound to a human copper chaperone (monomer) - new refinement | | Descriptor: | 3,3',3''-phosphanetriyltripropanoic acid, Copper transport protein ATOX1, PLATINUM (II) ION, ... | | Authors: | Shabalin, I.G, Boal, A.K, Dauter, Z, Jaskolski, M, Minor, W, Rosenzweig, A.C, Wlodawer, A. | | Deposit date: | 2015-02-23 | | Release date: | 2015-03-04 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.602 Å) | | Cite: | Crystallography and chemistry should always go together: a cautionary tale of protein complexes with cisplatin and carboplatin.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4YEA
 
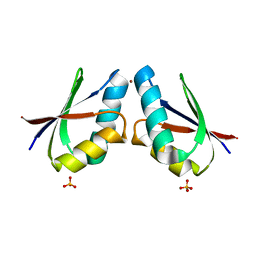 | | Crystal structure of cisplatin bound to a human copper chaperone (dimer) - new refinement | | Descriptor: | COPPER (II) ION, Copper transport protein ATOX1, SULFATE ION | | Authors: | Shabalin, I.G, Dauter, Z, Jaskolski, M, Minor, W, Wlodawer, A. | | Deposit date: | 2015-02-23 | | Release date: | 2015-03-18 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Crystallography and chemistry should always go together: a cautionary tale of protein complexes with cisplatin and carboplatin.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
1FAN
 
 | | CREVICE-FORMING MUTANTS IN THE RIGID CORE OF BOVINE PANCREATIC TRYPSIN INHIBITOR: CRYSTAL STRUCTURES OF F22A, Y23A, N43G, AND F45A | | Descriptor: | BOVINE PANCREATIC TRYPSIN INHIBITOR | | Authors: | Danishefsky, A.T, Wlodawer, A, Kim, K.-S, Tao, F, Woodward, C. | | Deposit date: | 1992-08-21 | | Release date: | 1993-10-31 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crevice-forming mutants in the rigid core of bovine pancreatic trypsin inhibitor: crystal structures of F22A, Y23A, N43G, and F45A.
Protein Sci., 2, 1993
|
|
1BTI
 
 | | CREVICE-FORMING MUTANTS IN THE RIGID CORE OF BOVINE PANCREATIC TRYPSIN INHIBITOR: CRYSTAL STRUCTURES OF F22A, Y23A, N43G, AND F45A | | Descriptor: | BOVINE PANCREATIC TRYPSIN INHIBITOR | | Authors: | Housset, D, Tao, F, Kim, K.-S, Fuchs, J, Woodward, C, Wlodawer, A. | | Deposit date: | 1991-07-11 | | Release date: | 1993-10-31 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crevice-forming mutants in the rigid core of bovine pancreatic trypsin inhibitor: crystal structures of F22A, Y23A, N43G, and F45A.
Protein Sci., 2, 1993
|
|
1BPT
 
 | | CREVICE-FORMING MUTANTS OF BPTI: CRYSTAL STRUCTURES OF F22A, Y23A, N43G, AND F45A | | Descriptor: | BOVINE PANCREATIC TRYPSIN INHIBITOR, PHOSPHATE ION | | Authors: | Housset, D, Wlodawer, A, Tao, F, Fuchs, J, Woodward, C. | | Deposit date: | 1991-12-11 | | Release date: | 1993-01-15 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crevice-forming mutants in the rigid core of bovine pancreatic trypsin inhibitor: crystal structures of F22A, Y23A, N43G, and F45A.
Protein Sci., 2, 1993
|
|
5C9D
 
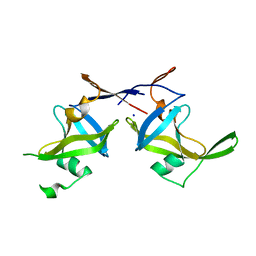 | | Crystal structure of a retropepsin-like aspartic protease from Rickettsia conorii | | Descriptor: | ApRick protease, SODIUM ION | | Authors: | Li, M, Gustchina, A, Cruz, R, Simoes, M, Curto, P, Martinez, J, Faro, C, Simoes, I, Wlodawer, A. | | Deposit date: | 2015-06-26 | | Release date: | 2015-10-14 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Structure of RC1339/APRc from Rickettsia conorii, a retropepsin-like aspartic protease
Acta Crystallogr.,Sect.D, 71, 2015
|
|
