7QQU
 
 | | SpCas9 bound to FANCF off-target2 DNA substrate | | Descriptor: | CRISPR-associated endonuclease Cas9/Csn1, FANCF off-target2 non-target strand, FANCF off-target2 target strand, ... | | Authors: | Pacesa, M, Jinek, M. | | Deposit date: | 2022-01-10 | | Release date: | 2022-10-26 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Structural basis for Cas9 off-target activity.
Cell, 185, 2022
|
|
7QQT
 
 | | SpCas9 bound to FANCF off-target1 DNA substrate | | Descriptor: | CRISPR-associated endonuclease Cas9/Csn1, FANCF off-target1 non-target strand, FANCF off-target1 target strand, ... | | Authors: | Pacesa, M, Jinek, M. | | Deposit date: | 2022-01-10 | | Release date: | 2022-10-26 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis for Cas9 off-target activity.
Cell, 185, 2022
|
|
7QR5
 
 | | SpCas9 bound to FANCF off-target6 DNA substrate | | Descriptor: | 1,2-ETHANEDIOL, CRISPR-associated endonuclease Cas9/Csn1, FANCF off-target6 non-target strand, ... | | Authors: | Pacesa, M, Jinek, M. | | Deposit date: | 2022-01-17 | | Release date: | 2022-10-26 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structural basis for Cas9 off-target activity.
Cell, 185, 2022
|
|
7QR8
 
 | | SpCas9 bound to PTPRC off-target1 DNA substrate | | Descriptor: | CRISPR-associated endonuclease Cas9/Csn1, MAGNESIUM ION, POTASSIUM ION, ... | | Authors: | Pacesa, M, Jinek, M. | | Deposit date: | 2022-01-10 | | Release date: | 2022-10-26 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structural basis for Cas9 off-target activity.
Cell, 185, 2022
|
|
7QQP
 
 | | SpCas9 bound to AAVS1 off-target3 DNA substrate | | Descriptor: | 1,2-ETHANEDIOL, AAVS1 off-target3 non-target strand, AAVS1 off-target3 target strand, ... | | Authors: | Pacesa, M, JInek, M. | | Deposit date: | 2022-01-10 | | Release date: | 2022-10-26 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural basis for Cas9 off-target activity.
Cell, 185, 2022
|
|
7QQS
 
 | | SpCas9 bound to FANCF on-target DNA substrate | | Descriptor: | CRISPR-associated endonuclease Cas9/Csn1, FANCF on-target non-target strand, FANCF on-target target strand, ... | | Authors: | Pacesa, M, Jinek, M. | | Deposit date: | 2022-01-10 | | Release date: | 2022-10-26 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis for Cas9 off-target activity.
Cell, 185, 2022
|
|
7QQX
 
 | | SpCas9 bound to FANCF off-target5 DNA substrate | | Descriptor: | CRISPR-associated endonuclease Cas9/Csn1, FANCF off-target5 non-target strand, FANCF off-target5 target strand, ... | | Authors: | Pacesa, M, Jinek, M. | | Deposit date: | 2022-01-10 | | Release date: | 2022-10-26 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis for Cas9 off-target activity.
Cell, 185, 2022
|
|
7QR0
 
 | | SpCas9 bound to TRAC off-target1 DNA substrate | | Descriptor: | CRISPR-associated endonuclease Cas9/Csn1, MAGNESIUM ION, POTASSIUM ION, ... | | Authors: | Pacesa, M, Jinek, M. | | Deposit date: | 2022-01-10 | | Release date: | 2022-10-26 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis for Cas9 off-target activity.
Cell, 185, 2022
|
|
7QQZ
 
 | | SpCas9 bound to FANCF off-target7 DNA substrate | | Descriptor: | CRISPR-associated endonuclease Cas9/Csn1, FANCF off-target7 non-target strand, FANCF off-target7 target strand, ... | | Authors: | Pacesa, M, Jinek, M. | | Deposit date: | 2022-01-10 | | Release date: | 2022-10-26 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural basis for Cas9 off-target activity.
Cell, 185, 2022
|
|
7QQW
 
 | | SpCas9 bound to FANCF off-target4 DNA substrate | | Descriptor: | CRISPR-associated endonuclease Cas9/Csn1, FANCF off-target4 non-target strand, FANCF off-target4 target strand, ... | | Authors: | Pacesa, M, Jinek, M. | | Deposit date: | 2022-01-10 | | Release date: | 2022-10-26 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural basis for Cas9 off-target activity.
Cell, 185, 2022
|
|
7QR1
 
 | | SpCas9 bound to TRAC off-target2 DNA substrate | | Descriptor: | CRISPR-associated endonuclease Cas9/Csn1, MAGNESIUM ION, POTASSIUM ION, ... | | Authors: | Pacesa, M, Jinek, M. | | Deposit date: | 2022-01-10 | | Release date: | 2022-10-26 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural basis for Cas9 off-target activity.
Cell, 185, 2022
|
|
7QR7
 
 | | SpCas9 bound to AAVS1 off-target2 DNA substrate | | Descriptor: | AAVS1 off-target2 non-target strand, AAVS1 off-target2 target strand, AAVS1 sgRNA, ... | | Authors: | Pacesa, M, JInek, M. | | Deposit date: | 2022-01-10 | | Release date: | 2022-10-26 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural basis for Cas9 off-target activity.
Cell, 185, 2022
|
|
3SIX
 
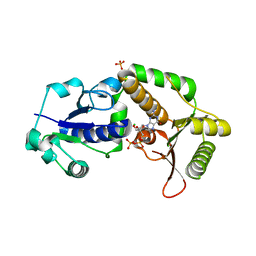 | | Crystal structure of NodZ alpha-1,6-fucosyltransferase soaked with GDP-fucose | | Descriptor: | CHLORIDE ION, GUANOSINE-5'-DIPHOSPHATE, Nodulation fucosyltransferase NodZ, ... | | Authors: | Brzezinski, K, Dauter, Z, Jaskolski, M. | | Deposit date: | 2011-06-20 | | Release date: | 2012-02-08 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structures of NodZ alpha-1,6-fucosyltransferase in complex with GDP and GDP-fucose
Acta Crystallogr.,Sect.D, 68, 2012
|
|
2ZXB
 
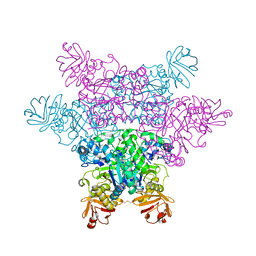 | | alpha-L-fucosidase complexed with inhibitor, ph-6FNJ | | Descriptor: | (2S,3R,4S,5R)-2-benzylpiperidine-3,4,5-triol, Alpha-L-fucosidase, putative | | Authors: | Wu, H.-J, Ko, T.-P, Ho, C.-W, Lin, C.-H, Wang, A.H.-J. | | Deposit date: | 2008-12-22 | | Release date: | 2009-12-08 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Structural basis of alpha-fucosidase inhibition by iminocyclitols with K(i) values in the micro- to picomolar range.
Angew.Chem.Int.Ed.Engl., 49, 2010
|
|
3SIW
 
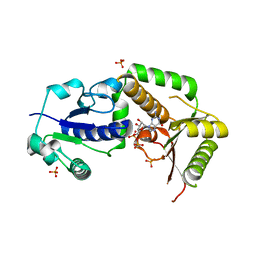 | | Crystal structure of NodZ alpha-1,6-fucosyltransferase co-crystallized with GDP | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, Nodulation fucosyltransferase NodZ, PHOSPHATE ION | | Authors: | Brzezinski, K, Dauter, Z, Jaskolski, M. | | Deposit date: | 2011-06-20 | | Release date: | 2012-02-08 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Structures of NodZ alpha-1,6-fucosyltransferase in complex with GDP and GDP-fucose
Acta Crystallogr.,Sect.D, 68, 2012
|
|
2ZXA
 
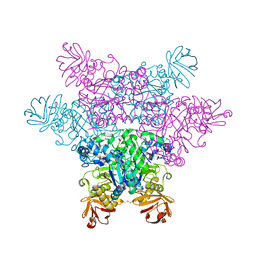 | | alpha-L-fucosidase complexed with inhibitor, FNJ-acetyl | | Descriptor: | Alpha-L-fucosidase, putative, N-{[(2R,3R,4R,5R,6S)-3,4,5-trihydroxy-6-methylpiperidin-2-yl]methyl}acetamide | | Authors: | Wu, H.-J, Ko, T.-P, Ho, C.-W, Lin, C.-H, Wang, A.H.-J. | | Deposit date: | 2008-12-22 | | Release date: | 2009-12-08 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.57 Å) | | Cite: | Structural basis of alpha-fucosidase inhibition by iminocyclitols with K(i) values in the micro- to picomolar range.
Angew.Chem.Int.Ed.Engl., 49, 2010
|
|
2ZX5
 
 | | alpha-L-fucosidase complexed with inhibitor, F10 | | Descriptor: | 3-(1H-indol-3-yl)-N-{[(2R,3R,4R,5R,6S)-3,4,5-trihydroxy-6-methylpiperidin-2-yl]methyl}propanamide, Alpha-L-fucosidase, putative | | Authors: | Wu, H.-J, Ko, T.-P, Ho, C.-W, Lin, C.-H, Wang, A.H.-J. | | Deposit date: | 2008-12-19 | | Release date: | 2009-12-08 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structural basis of alpha-fucosidase inhibition by iminocyclitols with K(i) values in the micro- to picomolar range.
Angew.Chem.Int.Ed.Engl., 49, 2010
|
|
2ZWZ
 
 | | alpha-L-fucosidase complexed with inhibitor, Core1 | | Descriptor: | (2R,3R,4R,5R,6S)-2-(aminomethyl)-6-methylpiperidine-3,4,5-triol, Alpha-L-fucosidase, putative | | Authors: | Wu, H.-J, Ko, T.-P, Ho, C.-W, Lin, C.-H, Wang, A.H.-J. | | Deposit date: | 2008-12-19 | | Release date: | 2009-12-08 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | Structural basis of alpha-fucosidase inhibition by iminocyclitols with K(i) values in the micro- to picomolar range.
Angew.Chem.Int.Ed.Engl., 49, 2010
|
|
2ZXD
 
 | | alpha-L-fucosidase complexed with inhibitor, iso-6FNJ | | Descriptor: | (2S,3R,4S,5R)-2-(1-methylethyl)piperidine-3,4,5-triol, Alpha-L-fucosidase, putative | | Authors: | Wu, H.-J, Ko, T.-P, Ho, C.-W, Lin, C.-H, Wang, A.H.-J. | | Deposit date: | 2008-12-22 | | Release date: | 2009-12-08 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structural basis of alpha-fucosidase inhibition by iminocyclitols with K(i) values in the micro- to picomolar range.
Angew.Chem.Int.Ed.Engl., 49, 2010
|
|
2ZX7
 
 | | alpha-L-fucosidase complexed with inhibitor, F10-2C | | Descriptor: | Alpha-L-fucosidase, putative, N-{[(2R,3R,4R,5R,6S)-3,4,5-trihydroxy-6-methylpiperidin-2-yl]methyl}-1H-indole-2-carboxamide | | Authors: | Wu, H.-J, Ko, T.-P, Ho, C.-W, Lin, C.-H, Wang, A.H.-J. | | Deposit date: | 2008-12-19 | | Release date: | 2009-12-08 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Structural basis of alpha-fucosidase inhibition by iminocyclitols with K(i) values in the micro- to picomolar range.
Angew.Chem.Int.Ed.Engl., 49, 2010
|
|
2ZX8
 
 | | alpha-L-fucosidase complexed with inhibitor, F10-2C-O | | Descriptor: | Alpha-L-fucosidase, putative, N-{[(2R,3R,4R,5R,6S)-3,4,5-trihydroxy-6-methylpiperidin-2-yl]methyl}-1-benzofuran-2-carboxamide | | Authors: | Wu, H.-J, Ko, T.-P, Ho, C.-W, Lin, C.-H, Wang, A.H.-J. | | Deposit date: | 2008-12-19 | | Release date: | 2009-12-08 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Structural basis of alpha-fucosidase inhibition by iminocyclitols with K(i) values in the micro- to picomolar range.
Angew.Chem.Int.Ed.Engl., 49, 2010
|
|
2ZX9
 
 | | alpha-L-fucosidase complexed with inhibitor, B4 | | Descriptor: | (2S)-2-cyclopentyl-2-phenyl-N-{[(2R,3R,4R,5R,6S)-3,4,5-trihydroxy-6-methylpiperidin-2-yl]methyl}ethanamide, Alpha-L-fucosidase, putative | | Authors: | Wu, H.-J, Ko, T.-P, Ho, C.-W, Lin, C.-H, Wang, A.H.-J. | | Deposit date: | 2008-12-22 | | Release date: | 2009-12-08 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.62 Å) | | Cite: | Structural basis of alpha-fucosidase inhibition by iminocyclitols with K(i) values in the micro- to picomolar range.
Angew.Chem.Int.Ed.Engl., 49, 2010
|
|
2ZX6
 
 | | alpha-L-fucosidase complexed with inhibitor, F10-1C | | Descriptor: | 2-(1H-indol-3-yl)-N-{[(2R,3R,4R,5R,6S)-3,4,5-trihydroxy-6-methylpiperidin-2-yl]methyl}acetamide, Alpha-L-fucosidase, putative | | Authors: | Wu, H.-J, Ko, T.-P, Ho, C.-W, Lin, C.-H, Wang, A.H.-J. | | Deposit date: | 2008-12-19 | | Release date: | 2009-12-08 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | Structural basis of alpha-fucosidase inhibition by iminocyclitols with K(i) values in the micro- to picomolar range.
Angew.Chem.Int.Ed.Engl., 49, 2010
|
|
3PJG
 
 | | Crystal structure of UDP-glucose dehydrogenase from Klebsiella pneumoniae complexed with product UDP-glucuronic acid | | Descriptor: | 3-CYCLOHEXYL-1-PROPYLSULFONIC ACID, UDP-glucose 6-dehydrogenase, URIDINE-5'-DIPHOSPHATE-GLUCURONIC ACID | | Authors: | Chen, Y.-Y, Ko, T.-P, Lin, C.-H, Chen, W.-H, Wang, A.H.-J. | | Deposit date: | 2010-11-10 | | Release date: | 2011-09-28 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Conformational change upon product binding to Klebsiella pneumoniae UDP-glucose dehydrogenase: a possible inhibition mechanism for the key enzyme in polymyxin resistance.
J.Struct.Biol., 175, 2011
|
|
3PLN
 
 | | Crystal structure of Klebsiella pneumoniae UDP-glucose 6-dehydrogenase complexed with UDP-glucose | | Descriptor: | UDP-glucose 6-dehydrogenase, URIDINE-5'-MONOPHOSPHATE | | Authors: | Chen, Y.-Y, Ko, T.-P, Lin, C.-H, Chen, W.-H, Wang, A.H.-J. | | Deposit date: | 2010-11-15 | | Release date: | 2011-09-28 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Conformational change upon product binding to Klebsiella pneumoniae UDP-glucose dehydrogenase: a possible inhibition mechanism for the key enzyme in polymyxin resistance.
J.Struct.Biol., 175, 2011
|
|
