1HR7
 
 | | Yeast Mitochondrial Processing Peptidase beta-E73Q Mutant | | Descriptor: | MITOCHONDRIAL PROCESSING PEPTIDASE ALPHA SUBUNIT, MITOCHONDRIAL PROCESSING PEPTIDASE BETA SUBUNIT, ZINC ION | | Authors: | Taylor, A.B, Smith, B.S, Kitada, S, Kojima, K, Miyaura, H, Otwinowski, Z, Ito, A, Deisenhofer, J. | | Deposit date: | 2000-12-21 | | Release date: | 2001-07-11 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Crystal structures of mitochondrial processing peptidase reveal the mode for specific cleavage of import signal sequences.
Structure, 9, 2001
|
|
1HR6
 
 | | Yeast Mitochondrial Processing Peptidase | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, MITOCHONDRIAL PROCESSING PEPTIDASE ALPHA SUBUNIT, MITOCHONDRIAL PROCESSING PEPTIDASE BETA SUBUNIT, ... | | Authors: | Taylor, A.B, Smith, B.S, Kitada, S, Kojima, K, Miyaura, H, Otwinowski, Z, Ito, A, Deisenhofer, J. | | Deposit date: | 2000-12-21 | | Release date: | 2001-07-11 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structures of mitochondrial processing peptidase reveal the mode for specific cleavage of import signal sequences.
Structure, 9, 2001
|
|
1HR8
 
 | | Yeast Mitochondrial Processing Peptidase beta-E73Q Mutant Complexed with Cytochrome C Oxidase IV Signal Peptide | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CYTOCHROME C OXIDASE POLYPEPTIDE IV, MITOCHONDRIAL PROCESSING PEPTIDASE ALPHA SUBUNIT, ... | | Authors: | Taylor, A.B, Smith, B.S, Kitada, S, Kojima, K, Miyaura, H, Otwinowski, Z, Ito, A, Deisenhofer, J. | | Deposit date: | 2000-12-21 | | Release date: | 2001-07-11 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structures of mitochondrial processing peptidase reveal the mode for specific cleavage of import signal sequences.
Structure, 9, 2001
|
|
2EJS
 
 | | Solution structure of RUH-076, a human CUE domain | | Descriptor: | Autocrine motility factor receptor, isoform 2 | | Authors: | Ruhul Momen, A.Z.M, Kitasaka, S, Hirota, H, Hayashi, F, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-03-20 | | Release date: | 2007-09-25 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of RUH-076, a human CUE domain
to be published
|
|
3AF8
 
 | | Crystal Structure of Pd(ally)/apo-C126AFr | | Descriptor: | 1,2-ETHANEDIOL, CADMIUM ION, Ferritin light chain, ... | | Authors: | Abe, S, Hikage, T, Watanabe, Y, Kitagawa, S, Ueno, T. | | Deposit date: | 2010-02-24 | | Release date: | 2010-08-11 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Mechanism of Accumulation and Incorporation of Organometallic Pd Complexes into the Protein Nanocage of apo-Ferritin.
Inorg.Chem., 49, 2010
|
|
3AF9
 
 | | Crystal Structure of Pd(allyl)/apo-C48AFr | | Descriptor: | 1,2-ETHANEDIOL, CADMIUM ION, Ferritin light chain, ... | | Authors: | Abe, S, Hikage, T, Watanabe, Y, Kitagawa, S, Ueno, T. | | Deposit date: | 2010-02-24 | | Release date: | 2010-08-11 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Mechanism of Accumulation and Incorporation of Organometallic Pd Complexes into the Protein Nanocage of apo-Ferritin.
Inorg.Chem., 49, 2010
|
|
3AF7
 
 | | Crystal Structure of 25Pd(allyl)/apo-Fr | | Descriptor: | 1,2-ETHANEDIOL, CADMIUM ION, Ferritin light chain, ... | | Authors: | Abe, S, Hikage, T, Watanabe, Y, Kitagawa, S, Ueno, T. | | Deposit date: | 2010-02-24 | | Release date: | 2010-08-11 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Mechanism of Accumulation and Incorporation of Organometallic Pd Complexes into the Protein Nanocage of apo-Ferritin.
Inorg.Chem., 49, 2010
|
|
3AZ4
 
 | | Crystal structure of Co/O-HEWL | | Descriptor: | CHLORIDE ION, COBALT (II) ION, Lysozyme C | | Authors: | Abe, S, Tsujimoto, M, Yoneda, K, Ohba, M, Hikage, T, Takano, M, Kitagawa, S, Ueno, T. | | Deposit date: | 2011-05-20 | | Release date: | 2012-05-09 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Porous protein crystals as reaction vessels for controlling magnetic properties of nanoparticles
Small, 8, 2012
|
|
3AZ5
 
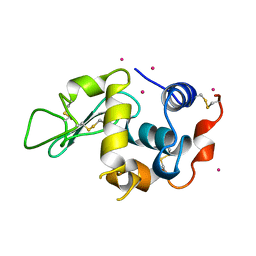 | | Crystal structure of Pt/O-HEWL | | Descriptor: | Lysozyme C, PLATINUM (II) ION | | Authors: | Abe, S, Tsujimoto, M, Yoneda, K, Ohba, M, Hikage, T, Takano, M, Kitagawa, S, Ueno, T. | | Deposit date: | 2011-05-20 | | Release date: | 2012-05-09 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Porous protein crystals as reaction vessels for controlling magnetic properties of nanoparticles
Small, 8, 2012
|
|
3AZ6
 
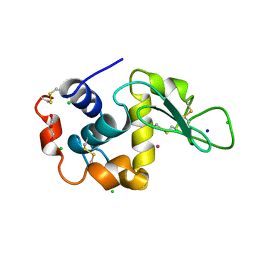 | | Crystal structure of Co/T-HEWL | | Descriptor: | CHLORIDE ION, COBALT (II) ION, GLYCEROL, ... | | Authors: | Abe, S, Tsujimoto, M, Yoneda, K, Ohba, M, Hikage, T, Takano, M, Kitagawa, S, Ueno, T. | | Deposit date: | 2011-05-20 | | Release date: | 2012-05-09 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Porous protein crystals as reaction vessels for controlling magnetic properties of nanoparticles
Small, 8, 2012
|
|
3AZ7
 
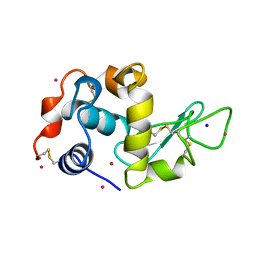 | | Crystal structure of Pt/T-HEWL | | Descriptor: | Lysozyme C, PLATINUM (II) ION, SODIUM ION | | Authors: | Abe, S, Tsujimoto, M, Yoneda, K, Ohba, M, Hikage, T, Takano, M, Kitagawa, S, Ueno, T. | | Deposit date: | 2011-05-20 | | Release date: | 2012-05-09 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Porous protein crystals as reaction vessels for controlling magnetic properties of nanoparticles
Small, 8, 2012
|
|
3ASE
 
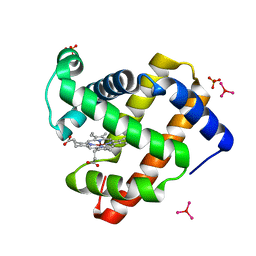 | | Crystal Structure of Zinc myoglobin soaked with Ru3O cluster | | Descriptor: | Myoglobin, PROTOPORPHYRIN IX CONTAINING ZN, SULFATE ION, ... | | Authors: | Koshiyama, T, Shirai, M, Hikage, T, Tabe, H, Tanaka, K, Kitagawa, S, Ueno, T. | | Deposit date: | 2010-12-11 | | Release date: | 2011-04-27 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Post-Crystal Engineering of Zinc-Substituted Myoglobin to Construct a Long-Lived Photoinduced Charge-Separation System
Angew.Chem.Int.Ed.Engl., 2011
|
|
3VKM
 
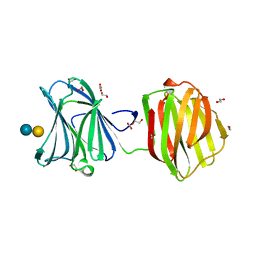 | | Protease-resistant mutant form of Human Galectin-8 in complex with sialyllactose and lactose | | Descriptor: | 1,2-ETHANEDIOL, Galectin-8, N-acetyl-alpha-neuraminic acid-(2-3)-beta-D-galactopyranose-(1-4)-beta-D-glucopyranose, ... | | Authors: | Yoshida, H, Yamashita, S, Teraoka, M, Nakakita, S, Nishi, N, Kamitori, S. | | Deposit date: | 2011-11-18 | | Release date: | 2012-09-12 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.98 Å) | | Cite: | X-ray structure of a protease-resistant mutant form of human galectin-8 with two carbohydrate recognition domains
Febs J., 279, 2012
|
|
3W6A
 
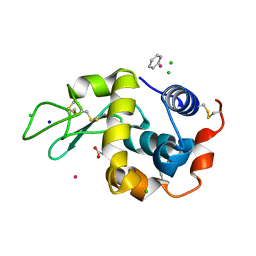 | | Crystal structure of cross-linked tetragonal hen egg white lysozyme soaked wiht 5mM [Ru(benzene)Cl2]2 | | Descriptor: | ACETATE ION, Benzeneruthenium(II) chloride, CHLORIDE ION, ... | | Authors: | Tabe, H, Abe, S, Hikage, T, Kitagawa, S, Ueno, T. | | Deposit date: | 2013-02-13 | | Release date: | 2014-02-19 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Solid Artificial Metalloenzymes: Post-Engineering of Porous Protein Crystals by Organometallic Complexes
To be Published
|
|
3WNL
 
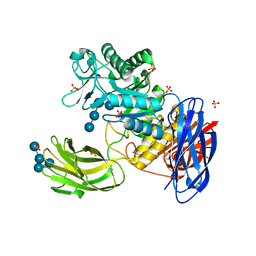 | | D308A mutant of Bacillus circulans T-3040 cycloisomaltooligosaccharide glucanotransferase complexed with isomaltohexaose | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, Cycloisomaltooligosaccharide glucanotransferase, ... | | Authors: | Suzuki, N, Fujimoto, Z, Kim, Y.M, Momma, M, Kishine, N, Suzuki, R, Suzuki, S, Kitamura, S, Kobayashi, M, Kimura, A, Funane, K. | | Deposit date: | 2013-12-10 | | Release date: | 2014-02-05 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural elucidation of the cyclization mechanism of alpha-1,6-glucan by Bacillus circulans T-3040 cycloisomaltooligosaccharide glucanotransferase.
J.Biol.Chem., 289, 2014
|
|
3WNN
 
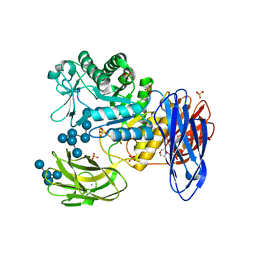 | | D308A mutant of Bacillus circulans T-3040 cycloisomaltooligosaccharide glucanotransferase complexed with isomaltooctaose | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, Cycloisomaltooligosaccharide glucanotransferase, ... | | Authors: | Suzuki, N, Fujimoto, Z, Kim, Y.M, Momma, M, Kishine, N, Suzuki, R, Suzuki, S, Kitamura, S, Kobayashi, M, Kimura, A, Funane, K. | | Deposit date: | 2013-12-10 | | Release date: | 2014-02-05 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural elucidation of the cyclization mechanism of alpha-1,6-glucan by Bacillus circulans T-3040 cycloisomaltooligosaccharide glucanotransferase.
J.Biol.Chem., 289, 2014
|
|
3WNM
 
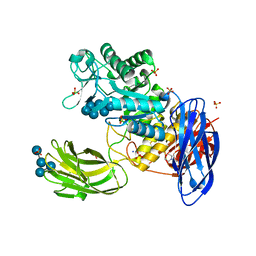 | | D308A mutant of Bacillus circulans T-3040 cycloisomaltooligosaccharide glucanotransferase complexed with isomaltoheptaose | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, Cycloisomaltooligosaccharide glucanotransferase, ... | | Authors: | Suzuki, N, Fujimoto, Z, Kim, Y.M, Momma, M, Kishine, N, Suzuki, R, Suzuki, S, Kitamura, S, Kobayashi, M, Kimura, A, Funane, K. | | Deposit date: | 2013-12-10 | | Release date: | 2014-02-05 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural elucidation of the cyclization mechanism of alpha-1,6-glucan by Bacillus circulans T-3040 cycloisomaltooligosaccharide glucanotransferase.
J.Biol.Chem., 289, 2014
|
|
3WNP
 
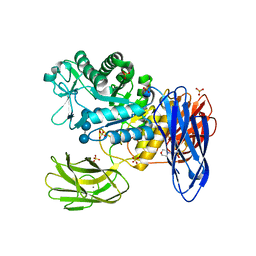 | | D308A, F268V, D469Y, A513V, and Y515S quintuple mutant of Bacillus circulans T-3040 cycloisomaltooligosaccharide glucanotransferase complexed with isomaltoundecaose | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, Cycloisomaltooligosaccharide glucanotransferase, ... | | Authors: | Suzuki, R, Suzuki, N, Fujimoto, Z, Momma, M, Kimura, K, Kitamura, S, Kimura, A, Funane, K. | | Deposit date: | 2013-12-10 | | Release date: | 2014-02-05 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Molecular engineering of cycloisomaltooligosaccharide glucanotransferase from Bacillus circulans T-3040: structural determinants for the reaction product size and reactivity.
Biochem.J., 467, 2015
|
|
8SD4
 
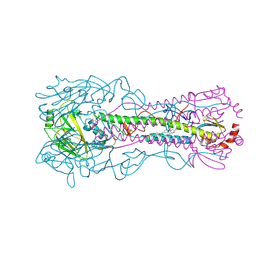 | | Crystal structure of the A/Puerto Rico/8/1934 (H1N1) influenza virus hemagglutinin in complex with small molecule fusion inhibitor compound 7 | | Descriptor: | (S~1~S)-N-{3,5-dichloro-4-[(2S)-2-phenylmorpholine-4-carbonyl]phenyl}-3-[(dimethylamino)methyl]azetidine-1-sulfonimidoyl fluoride, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin HA1 chain, ... | | Authors: | Kadam, R.U, Zhu, X, Wilson, I.A. | | Deposit date: | 2023-04-06 | | Release date: | 2024-06-05 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.11 Å) | | Cite: | Ultrapotent influenza hemagglutinin fusion inhibitors developed through SuFEx-enabled high-throughput medicinal chemistry.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8SD2
 
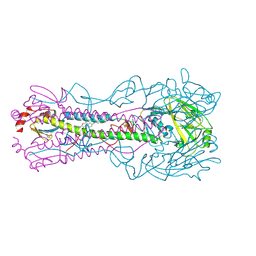 | | Crystal structure of the A/Puerto Rico/8/1934 (H1N1) influenza virus hemagglutinin in complex with small molecule fusion inhibitor compound 4 | | Descriptor: | (S~6~S)-N-{3-chloro-4-[(2S)-2-phenylmorpholine-4-carbonyl]phenyl}-5,7-dihydro-6H-pyrrolo[3,4-b]pyridine-6-sulfonimidoyl fluoride, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Kadam, R.U, Zhu, X.Y, Wilson, I.A. | | Deposit date: | 2023-04-06 | | Release date: | 2024-06-05 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | Ultrapotent influenza hemagglutinin fusion inhibitors developed through SuFEx-enabled high-throughput medicinal chemistry.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8VQQ
 
 | | Crystal structure of the A/Puerto Rico/8/1934 (H1N1) influenza virus hemagglutinin in complex with small molecule 6S | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin HA1 chain, Hemagglutinin HA2 chain, ... | | Authors: | Lin, T.H, Zhu, Y, Wilson, I.A. | | Deposit date: | 2024-01-19 | | Release date: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Ultrapotent influenza hemagglutinin fusion inhibitors developed through SuFEx-enabled high-throughput medicinal chemistry.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8VQM
 
 | | Crystal structure of the A/Puerto Rico/8/1934 (H1N1) influenza virus hemagglutinin in complex with small molecule 6R prime | | Descriptor: | (S~1~S,3R)-N-{3,5-dichloro-4-[(2S)-2-phenylmorpholine-4-carbonyl]phenyl}-3-(dimethylamino)pyrrolidine-1-sulfonimidoyl fluoride, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Lin, T.H, Zhu, Y, Wilson, I.A. | | Deposit date: | 2024-01-18 | | Release date: | 2024-06-05 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Ultrapotent influenza hemagglutinin fusion inhibitors developed through SuFEx-enabled high-throughput medicinal chemistry.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8VQN
 
 | | Crystal structure of the A/Puerto Rico/8/1934 (H1N1) influenza virus hemagglutinin in complex with small molecule 6R | | Descriptor: | (S~1~S,3R)-N-{3-chloro-4-[(2S)-2-phenylmorpholine-4-carbonyl]phenyl}-3-(dimethylamino)pyrrolidine-1-sulfonimidoyl fluoride, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin HA1 chain, ... | | Authors: | Lin, T.H, Zhu, Y, Wilson, I.A. | | Deposit date: | 2024-01-18 | | Release date: | 2024-06-05 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Ultrapotent influenza hemagglutinin fusion inhibitors developed through SuFEx-enabled high-throughput medicinal chemistry.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8VQL
 
 | | Crystal structure of the A/Puerto Rico/8/1934 (H1N1) influenza virus hemagglutinin in complex with small molecule 6S prime | | Descriptor: | (S~1~S,3S)-N-{3,5-dichloro-4-[(2S)-2-phenylmorpholine-4-carbonyl]phenyl}-3-(dimethylamino)pyrrolidine-1-sulfonimidoyl fluoride, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Lin, T.H, Zhu, Y, Wilson, I.A. | | Deposit date: | 2024-01-18 | | Release date: | 2024-06-05 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Ultrapotent influenza hemagglutinin fusion inhibitors developed through SuFEx-enabled high-throughput medicinal chemistry.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
5HK9
 
 | |
