3E5K
 
 | | Crystal structure of CYP105P1 wild-type 4-phenylimidazole complex | | Descriptor: | 4-PHENYL-1H-IMIDAZOLE, Cytochrome P450 (Cytochrome P450 hydroxylase), PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Xu, L.H, Fushinobu, S, Ikeda, H, Wakagi, T, Shoun, H. | | Deposit date: | 2008-08-14 | | Release date: | 2008-12-30 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structures of cytochrome P450 105P1 from Streptomyces avermitilis: conformational flexibility and histidine ligation state
J.Bacteriol., 191, 2009
|
|
3EQN
 
 | | Crystal structure of beta-1,3-glucanase from Phanerochaete chrysosporium (Lam55A) | | Descriptor: | ACETATE ION, GLYCEROL, Glucan 1,3-beta-glucosidase, ... | | Authors: | Ishida, T, Fushinobu, S, Kawai, R, Kitaoka, M, Igarashi, K, Samejima, M. | | Deposit date: | 2008-10-01 | | Release date: | 2009-02-03 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of glycoside hydrolase family 55 beta -1,3-glucanase from the basidiomycete Phanerochaete chrysosporium
J.Biol.Chem., 284, 2009
|
|
3E5L
 
 | | Crystal structure of CYP105P1 H72A mutant | | Descriptor: | Cytochrome P450 (Cytochrome P450 hydroxylase), PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Xu, L.H, Fushinobu, S, Ikeda, H, Wakagi, T, Shoun, H. | | Deposit date: | 2008-08-14 | | Release date: | 2008-12-30 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structures of cytochrome P450 105P1 from Streptomyces avermitilis: conformational flexibility and histidine ligation state
J.Bacteriol., 191, 2009
|
|
3EQO
 
 | | Crystal structure of beta-1,3-glucanase from Phanerochaete chrysosporium (Lam55A) gluconolactone complex | | Descriptor: | D-glucono-1,5-lactone, Glucan 1,3-beta-glucosidase, ZINC ION, ... | | Authors: | Ishida, T, Fushinobu, S, Kawai, R, Kitaoka, M, Igarashi, K, Samejima, M. | | Deposit date: | 2008-10-01 | | Release date: | 2009-02-03 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Crystal structure of glycoside hydrolase family 55 beta -1,3-glucanase from the basidiomycete Phanerochaete chrysosporium
J.Biol.Chem., 284, 2009
|
|
6KXE
 
 | | The ishigamide ketosynthase/chain length factor | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, Ketosynthase, ... | | Authors: | Du, D, Katsuyama, Y, Horiuchi, M, Fushinobu, S, Chen, A, Davis, T, Burkart, M, Ohnishi, Y. | | Deposit date: | 2019-09-10 | | Release date: | 2020-05-06 | | Last modified: | 2025-04-09 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Structural basis for selectivity in a highly reducing type II polyketide synthase.
Nat.Chem.Biol., 16, 2020
|
|
6KXF
 
 | | The ishigamide ketosynthase/chain length factor | | Descriptor: | ACP, Ketosynthase, [(3~{R})-2,2-dimethyl-4-[[3-[2-[[(~{E})-oct-2-enoyl]amino]ethylamino]-3-oxidanylidene-propyl]amino]-3-oxidanyl-4-oxidanylidene-butyl] dihydrogen phosphate | | Authors: | Du, D, Katsuyama, Y, Horiuchi, M, Fushinobu, S, Chen, A, Davis, T, Burkart, M, Ohnishi, Y. | | Deposit date: | 2019-09-10 | | Release date: | 2020-05-06 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Structural basis for selectivity in a highly reducing type II polyketide synthase.
Nat.Chem.Biol., 16, 2020
|
|
6KXD
 
 | | The ishigamide ketosynthase/chain length factor | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, Ketosynthase, ... | | Authors: | Du, D, Katsuyama, Y, Horiuchi, M, Fushinobu, S, Chen, A, Davis, T, Burkart, M, Ohnishi, Y. | | Deposit date: | 2019-09-10 | | Release date: | 2020-05-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural basis for selectivity in a highly reducing type II polyketide synthase.
Nat.Chem.Biol., 16, 2020
|
|
8IC1
 
 | | endo-alpha-D-arabinanase EndoMA1 D51N mutant from Microbacterium arabinogalactanolyticum in complex with arabinooligosaccharides | | Descriptor: | (3~{a}~{S},5~{R},6~{R},6~{a}~{S})-5-(hydroxymethyl)-2,2-dimethyl-3~{a},5,6,6~{a}-tetrahydrofuro[2,3-d][1,3]dioxol-6-ol, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, ... | | Authors: | Li, J, Nakashima, C, Ishiwata, A, Fujita, K, Fushinobu, S. | | Deposit date: | 2023-02-10 | | Release date: | 2023-08-16 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Identification and characterization of endo-alpha-, exo-alpha-, and exo-beta-D-arabinofuranosidases degrading lipoarabinomannan and arabinogalactan of mycobacteria.
Nat Commun, 14, 2023
|
|
8IC8
 
 | | Exo-alpha-D-arabinofuranosidase from Microbacterium arabinogalactanolyticum | | Descriptor: | Exo-alpha-D-arabinofuranosidase, PHOSPHATE ION | | Authors: | Kashima, T, Arakawa, T, Yamada, C, Ishiwata, A, Fujita, K, Fushinobu, S. | | Deposit date: | 2023-02-11 | | Release date: | 2023-08-16 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | Identification and characterization of endo-alpha-, exo-alpha-, and exo-beta-D-arabinofuranosidases degrading lipoarabinomannan and arabinogalactan of mycobacteria.
Nat Commun, 14, 2023
|
|
8IC7
 
 | | exo-beta-D-arabinofuranosidase ExoMA2 from Microbacterium arabinogalactanolyticum in complex with beta-D-arabinofuranose | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Fukushima, R, Kashima, T, Ishiwata, A, Fujita, K, Fushinobu, S. | | Deposit date: | 2023-02-11 | | Release date: | 2023-08-16 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Identification and characterization of endo-alpha-, exo-alpha-, and exo-beta-D-arabinofuranosidases degrading lipoarabinomannan and arabinogalactan of mycobacteria.
Nat Commun, 14, 2023
|
|
8IC6
 
 | | exo-beta-D-arabinanase ExoMA2 from Microbacterium arabinogalactanolyticum in complex with Tris | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, ... | | Authors: | Fukushima, R, Kashima, T, Ishiwata, A, Fujita, K, Fushinobu, S. | | Deposit date: | 2023-02-10 | | Release date: | 2023-08-16 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Identification and characterization of endo-alpha-, exo-alpha-, and exo-beta-D-arabinofuranosidases degrading lipoarabinomannan and arabinogalactan of mycobacteria.
Nat Commun, 14, 2023
|
|
8HHV
 
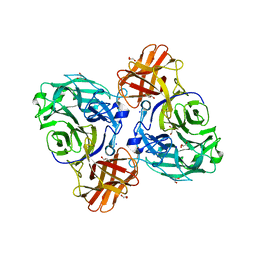 | | endo-alpha-D-arabinanase EndoMA1 from Microbacterium arabinogalactanolyticum | | Descriptor: | CALCIUM ION, GLYCEROL, SODIUM ION, ... | | Authors: | Nakashima, C, Li, J, Arakawa, T, Yamada, C, Ishiwata, A, Fujita, K, Fushinobu, S. | | Deposit date: | 2022-11-17 | | Release date: | 2023-08-16 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Identification and characterization of endo-alpha-, exo-alpha-, and exo-beta-D-arabinofuranosidases degrading lipoarabinomannan and arabinogalactan of mycobacteria.
Nat Commun, 14, 2023
|
|
6LCE
 
 | |
6L69
 
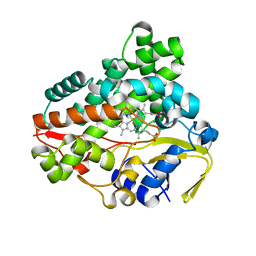 | | Crystal structure of CYP154C2 from Streptomyces avermitilis | | Descriptor: | Cytochrome P450 hydroxylase, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Xu, L.H, Fushinobu, S. | | Deposit date: | 2019-10-28 | | Release date: | 2020-09-23 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Regio- and stereoselective hydroxylation of testosterone by a novel cytochrome P450 154C2 from Streptomyces avermitilis.
Biochem.Biophys.Res.Commun., 522, 2020
|
|
6LCF
 
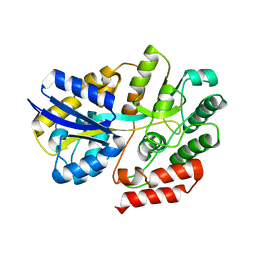 | | Crystal Structure of beta-L-arabinobiose binding protein - native | | Descriptor: | ABC transporter substrate binding component, beta-L-arabinofuranose-(1-2)-beta-L-arabinofuranose | | Authors: | Miyake, M, Arakawa, T, Fushinobu, S. | | Deposit date: | 2019-11-18 | | Release date: | 2020-04-22 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Structural analysis of beta-L-arabinobiose-binding protein in the metabolic pathway of hydroxyproline-rich glycoproteins in Bifidobacterium longum.
Febs J., 287, 2020
|
|
8K2N
 
 | |
8K2J
 
 | |
8K2K
 
 | | Crystal structure of Group 3 Oligosaccharide/Monosaccharide-releasing beta-N-acetylgalactosaminidase NgaDssm in complex with GalNAc-thiazoline | | Descriptor: | (3aR,5R,6R,7R,7aR)-5-(hydroxymethyl)-2-methyl-5,6,7,7a-tetrahydro-3aH-pyrano[3,2-d][1,3]thiazole-6,7-diol, CHLORIDE ION, GLYCEROL, ... | | Authors: | Sumida, T, Fushinobu, S. | | Deposit date: | 2023-07-12 | | Release date: | 2024-04-24 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Genetic and functional diversity of beta-N-acetylgalactosamine-targeting glycosidases expanded by deep-sea metagenome analysis.
Nat Commun, 15, 2024
|
|
8K2H
 
 | | Crystal structure of Group 2Oligosaccharide/Monosaccharide-releasing beta-N-acetylhexosaminidase NgaAt from Arabidopsis thaliana in complex with GalNAc-thiazoline | | Descriptor: | (3aR,5R,6R,7R,7aR)-5-(hydroxymethyl)-2-methyl-5,6,7,7a-tetrahydro-3aH-pyrano[3,2-d][1,3]thiazole-6,7-diol, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Sumida, T, Fushinobu, S. | | Deposit date: | 2023-07-12 | | Release date: | 2024-04-24 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Genetic and functional diversity of beta-N-acetylgalactosamine-targeting glycosidases expanded by deep-sea metagenome analysis.
Nat Commun, 15, 2024
|
|
8K2F
 
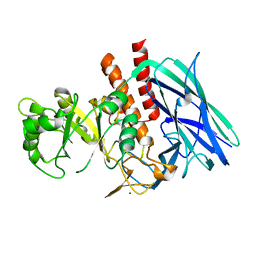 | |
8K2I
 
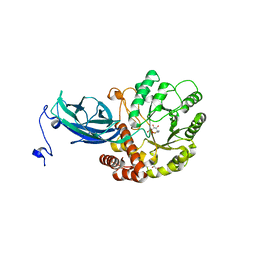 | | Crystal structure of Group 2 Oligosaccharide/Monosaccharide-releasing beta-N-acetylhexosaminidase NgaAt from Arabidopsis thaliana in complex with GlcNAc-thiazoline | | Descriptor: | 3AR,5R,6S,7R,7AR-5-HYDROXYMETHYL-2-METHYL-5,6,7,7A-TETRAHYDRO-3AH-PYRANO[3,2-D]THIAZOLE-6,7-DIOL, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Sumida, T, Fushinobu, S. | | Deposit date: | 2023-07-12 | | Release date: | 2024-04-24 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Genetic and functional diversity of beta-N-acetylgalactosamine-targeting glycosidases expanded by deep-sea metagenome analysis.
Nat Commun, 15, 2024
|
|
8K2M
 
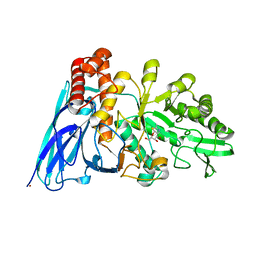 | | Crystal structure of Group 4 Monosaccharide-releasing beta-N-acetylgalactosaminidase NgaP2 from Paenibacillus sp. TS12 in complex with GalNAc-thiazoline | | Descriptor: | (3aR,5R,6R,7R,7aR)-5-(hydroxymethyl)-2-methyl-5,6,7,7a-tetrahydro-3aH-pyrano[3,2-d][1,3]thiazole-6,7-diol, BROMIDE ION, Monosaccharide-releasing beta-N-acetylgalactosaminidase | | Authors: | Sumida, T, Fushinobu, S. | | Deposit date: | 2023-07-12 | | Release date: | 2024-04-24 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Genetic and functional diversity of beta-N-acetylgalactosamine-targeting glycosidases expanded by deep-sea metagenome analysis.
Nat Commun, 15, 2024
|
|
8K2L
 
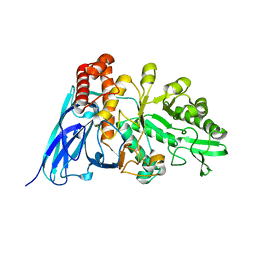 | |
8K2G
 
 | |
6M3X
 
 | | Cryo-EM structure of sulfur oxygenase reductase from Sulfurisphaera tokodaii | | Descriptor: | FE (III) ION, Sulfur oxygenase/reductase | | Authors: | Sato, Y, Adachi, N, Moriya, T, Arakawa, T, Kawasaki, M, Yamada, C, Senda, T, Fushinobu, S. | | Deposit date: | 2020-03-04 | | Release date: | 2020-07-15 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (2.24 Å) | | Cite: | Crystallographic and cryogenic electron microscopic structures and enzymatic characterization of sulfur oxygenase reductase fromSulfurisphaera tokodaii.
J Struct Biol X, 4, 2020
|
|
