5XW6
 
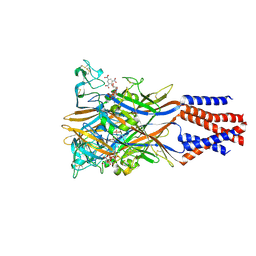 | | Crystal structure of the chicken ATP-gated P2X7 receptor channel in the presence of competitive antagonist TNP-ATP at 3.1 Angstroms | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, P2X purinoceptor, SPIRO(2,4,6-TRINITROBENZENE[1,2A]-2O',3O'-METHYLENE-ADENINE-TRIPHOSPHATE | | Authors: | Kasuya, G, Hattori, M, Nureki, O. | | Deposit date: | 2017-06-29 | | Release date: | 2017-10-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural insights into the competitive inhibition of the ATP-gated P2X receptor channel
Nat Commun, 8, 2017
|
|
5XVR
 
 | |
5Y50
 
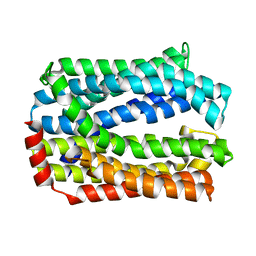 | | Crystal structure of eukaryotic MATE transporter AtDTX14 | | Descriptor: | Protein DETOXIFICATION 14 | | Authors: | Miyauchi, H, Kusakizako, T, Nishizawa, T, Ishitani, R, Nureki, O. | | Deposit date: | 2017-08-06 | | Release date: | 2017-12-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural basis for xenobiotic extrusion by eukaryotic MATE transporter
Nat Commun, 8, 2017
|
|
5ZHS
 
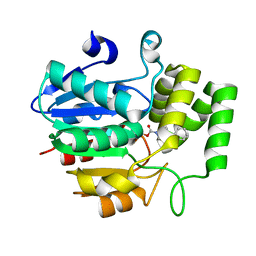 | | Crystal structure of OsD14 in complex with covalently bound KK052 | | Descriptor: | (4-phenylpiperazin-1-yl)(1H-1,2,3-triazol-1-yl)methanone, Strigolactone esterase D14 | | Authors: | Hirabayashi, K, Miyakawa, T, Tanokura, M. | | Deposit date: | 2018-03-13 | | Release date: | 2018-11-21 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Triazole Ureas Covalently Bind to Strigolactone Receptor and Antagonize Strigolactone Responses.
Mol Plant, 12, 2019
|
|
5ZHT
 
 | | Crystal structure of OsD14 in complex with covalently bound KK073 | | Descriptor: | (1H-1,2,3-triazol-1-yl){4-[4-(trifluoromethyl)phenyl]piperazin-1-yl}methanone, Strigolactone esterase D14 | | Authors: | Hirabayashi, K, Miyakawa, T, Tanokura, M. | | Deposit date: | 2018-03-13 | | Release date: | 2018-11-21 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.532 Å) | | Cite: | Triazole Ureas Covalently Bind to Strigolactone Receptor and Antagonize Strigolactone Responses.
Mol Plant, 12, 2019
|
|
5WXI
 
 | | EarP bound with dTDP-rhamnose (soaked) | | Descriptor: | 2'-DEOXY-THYMIDINE-BETA-L-RHAMNOSE, BETA-MERCAPTOETHANOL, EarP, ... | | Authors: | Sengoku, T, Yokoyama, S, Yanagisawa, T. | | Deposit date: | 2017-01-07 | | Release date: | 2018-02-28 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis of protein arginine rhamnosylation by glycosyltransferase EarP
Nat. Chem. Biol., 14, 2018
|
|
5WXJ
 
 | | Apo EarP | | Descriptor: | BETA-MERCAPTOETHANOL, EarP, GLYCEROL, ... | | Authors: | Sengoku, T, Yokoyama, S, Yanagisawa, T. | | Deposit date: | 2017-01-07 | | Release date: | 2018-02-28 | | Last modified: | 2018-04-04 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural basis of protein arginine rhamnosylation by glycosyltransferase EarP
Nat. Chem. Biol., 14, 2018
|
|
5WXK
 
 | | EarP bound with domain I of EF-P | | Descriptor: | BETA-MERCAPTOETHANOL, EarP, Elongation factor P, ... | | Authors: | Sengoku, T, Yokoyama, S, Yanagisawa, T. | | Deposit date: | 2017-01-07 | | Release date: | 2018-02-28 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.801 Å) | | Cite: | Structural basis of protein arginine rhamnosylation by glycosyltransferase EarP
Nat. Chem. Biol., 14, 2018
|
|
5ZHR
 
 | | Crystal structure of OsD14 in complex with covalently bound KK094 | | Descriptor: | (2,3-dihydro-1H-indol-1-yl)(1H-1,2,3-triazol-1-yl)methanone, Strigolactone esterase D14 | | Authors: | Hirabayashi, K, Miyakawa, T, Tanokura, M. | | Deposit date: | 2018-03-13 | | Release date: | 2018-11-21 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Triazole Ureas Covalently Bind to Strigolactone Receptor and Antagonize Strigolactone Responses.
Mol Plant, 12, 2019
|
|
3X20
 
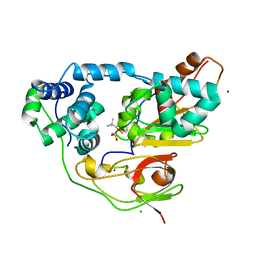 | | Crystal structure of Nitrile Hydratase mutant bR56K complexed with Trimethylacetonitrile, photo-activated for 25 min | | Descriptor: | 2,2-dimethylpropanenitrile, CHLORIDE ION, FE (III) ION, ... | | Authors: | Yamanaka, Y, Hashimoto, K, Noguchi, N, Yohda, M, Odaka, M. | | Deposit date: | 2014-12-03 | | Release date: | 2016-01-27 | | Method: | X-RAY DIFFRACTION (1.18 Å) | | Cite: | Time-Resolved Crystallography of the Reaction Intermediate of Nitrile Hydratase: Revealing a Role for the Cysteinesulfenic Acid Ligand as a Catalytic Nucleophile.
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
3A3G
 
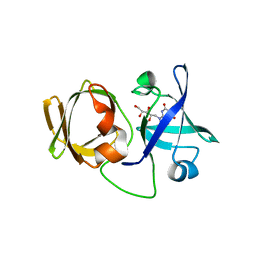 | | Crystal structure of LumP complexed with 6,7-dimethyl-8-(1'-D-ribityl) lumazine | | Descriptor: | 1-deoxy-1-(6,7-dimethyl-2,4-dioxo-3,4-dihydropteridin-8(2H)-yl)-D-ribitol, Lumazine protein | | Authors: | Sato, Y. | | Deposit date: | 2009-06-12 | | Release date: | 2009-11-10 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structures of the lumazine protein from Photobacterium kishitanii in complexes with the authentic chromophore, 6,7-dimethyl-8-(1'-D-ribityl) lumazine and its analogues, riboflavin and FMN, at high resolution
J.Bacteriol., 192, 2009
|
|
3A3B
 
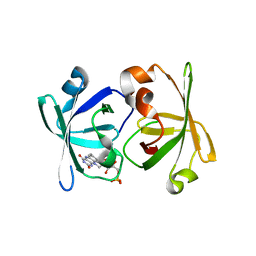 | | Crystal structure of LumP complexed with flavin mononucleotide | | Descriptor: | FLAVIN MONONUCLEOTIDE, Lumazine protein, RIBOFLAVIN | | Authors: | Sato, Y. | | Deposit date: | 2009-06-11 | | Release date: | 2009-11-10 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structures of the lumazine protein from Photobacterium kishitanii in complexes with the authentic chromophore, 6,7-dimethyl-8-(1'-D-ribityl) lumazine and its analogues, riboflavin and FMN, at high resolution
J.Bacteriol., 192, 2009
|
|
3A1K
 
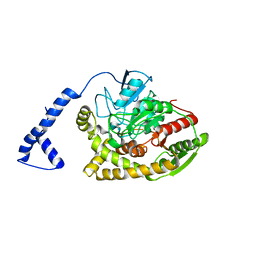 | | Crystal structure of Rhodococcus sp. N771 Amidase | | Descriptor: | Amidase | | Authors: | Ohtaki, A, Noguchi, K, Sato, Y, Murata, K, Odaka, M, Yohda, M. | | Deposit date: | 2009-04-09 | | Release date: | 2009-11-03 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Structure and characterization of amidase from Rhodococcus sp. N-771: Insight into the molecular mechanism of substrate recognition
Biochim.Biophys.Acta, 1804, 2010
|
|
3AEI
 
 | | Crystal structure of the prefoldin beta2 subunit from Thermococcus strain KS-1 | | Descriptor: | CHLORIDE ION, Prefoldin beta subunit 2, SULFATE ION | | Authors: | Ohtaki, A, Sugano, Y, Sato, T, Noguchi, K, Miyatake, H, Yohda, M. | | Deposit date: | 2010-02-08 | | Release date: | 2010-05-19 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Thermodynamic Characterization of the Interaction between Prefoldin and Group II Chaperonin
J.Mol.Biol., 399, 2010
|
|
3A1I
 
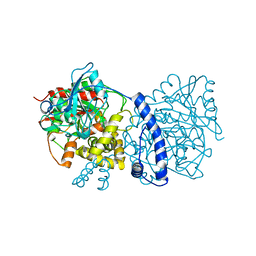 | | Crystal structure of Rhodococcus sp. N-771 Amidase complexed with Benzamide | | Descriptor: | Amidase, BENZAMIDE | | Authors: | Ohtaki, A, Noguchi, K, Sato, Y, Murata, K, Odaka, M, Yohda, M. | | Deposit date: | 2009-04-03 | | Release date: | 2009-10-27 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | Structure and Characterization of Amidase from Rhodococcus sp. N-771: Insight into the Molecular Mechanism of Substrate Recognition
Biochim.Biophys.Acta, 2009
|
|
3AY5
 
 | |
2ZJS
 
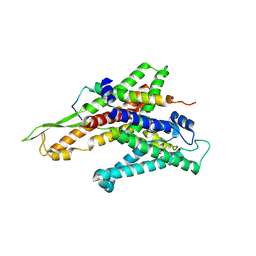 | | Crystal Structure of SecYE translocon from Thermus thermophilus with a Fab fragment | | Descriptor: | Fab56 (heavy chain), Fab56 (light chain), Preprotein translocase SecE subunit, ... | | Authors: | Tsukazaki, T, Mori, H, Fukai, S, Ishitani, R, Perederina, A, Vassylyev, D.G, Ito, K, Nureki, O. | | Deposit date: | 2008-03-08 | | Release date: | 2008-10-14 | | Last modified: | 2021-11-10 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Conformational transition of Sec machinery inferred from bacterial SecYE structures
Nature, 455, 2008
|
|
2ZQP
 
 | |
2ZZD
 
 | | Recombinant thiocyanate hydrolase, air-oxidized form of holo-enzyme | | Descriptor: | COBALT (III) ION, L(+)-TARTARIC ACID, Thiocyanate hydrolase subunit alpha, ... | | Authors: | Arakawa, T, Kawano, Y, Katayama, Y, Yohda, M, Odaka, M. | | Deposit date: | 2009-02-09 | | Release date: | 2009-10-13 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Structural Basis for Catalytic Activation of Thiocyanate Hydrolase Involving Metal-Ligated Cysteine Modification
J.Am.Chem.Soc., 131, 2009
|
|
3A35
 
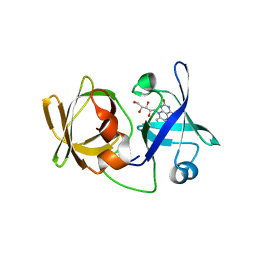 | | Crystal structure of LumP complexed with riboflavin | | Descriptor: | Lumazine protein, RIBOFLAVIN | | Authors: | Sato, Y. | | Deposit date: | 2009-06-09 | | Release date: | 2009-11-10 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.421 Å) | | Cite: | Crystal structures of the lumazine protein from Photobacterium kishitanii in complexes with the authentic chromophore, 6,7-dimethyl-8-(1'-D-ribityl) lumazine and its analogues, riboflavin and FMN, at high resolution
J.Bacteriol., 192, 2009
|
|
3VWA
 
 | | Crystal structure of Cex1p | | Descriptor: | Cytoplasmic export protein 1 | | Authors: | Nozawa, K, Ishitani, R, Nureki, O. | | Deposit date: | 2012-08-13 | | Release date: | 2013-06-05 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of Cex1p reveals the mechanism of tRNA trafficking between nucleus and cytoplasm
Nucleic Acids Res., 41, 2013
|
|
3UFZ
 
 | | Crystal structure of a Trp-less green fluorescent protein translated by the universal genetic code | | Descriptor: | Green fluorescent protein | | Authors: | Kawahara-Kobayashi, A, Araiso, Y, Matsuda, T, Yokoyama, S, Kigawa, T, Nureki, O, Kiga, D. | | Deposit date: | 2011-11-02 | | Release date: | 2012-10-17 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Simplification of the genetic code: restricted diversity of genetically encoded amino acids.
Nucleic Acids Res., 40, 2012
|
|
3VQJ
 
 | | Crystal Structutre of Thiobacillus thioparus THI115 Carbonyl Sulfide Hydrolase | | Descriptor: | Carbonyl sulfide hydrolase, SODIUM ION, ZINC ION | | Authors: | Katayama, Y, Noguchi, K, Ogawa, T, Ohtaki, A, Odaka, M, Yohda, M. | | Deposit date: | 2012-03-24 | | Release date: | 2013-02-27 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Carbonyl Sulfide Hydrolase from Thiobacillus thioparus Strain THI115 Is One of the beta-Carbonic Anhydrase Family Enzymes
J.Am.Chem.Soc., 135, 2013
|
|
3VRK
 
 | | Crystal Structutre of Thiobacillus thioparus THI115 Carbonyl Sulfide Hydrolase / Thiocyanate complex | | Descriptor: | Carbonyl sulfide hydrolase, SODIUM ION, THIOCYANATE ION, ... | | Authors: | Katayama, Y, Noguchi, K, Ogawa, T, Ohtaki, A, Odaka, M, Yohda, M. | | Deposit date: | 2012-04-11 | | Release date: | 2013-02-27 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.33 Å) | | Cite: | Carbonyl Sulfide Hydrolase from Thiobacillus thioparus Strain THI115 Is One of the beta-Carbonic Anhydrase Family Enzymes
J.Am.Chem.Soc., 135, 2013
|
|
3X24
 
 | | Crystal structure of Nitrile Hydratase mutant bR56K complexed with Trimethylacetonitrile, photo-activated for 120 min | | Descriptor: | 2,2-dimethylpropanenitrile, FE (III) ION, MAGNESIUM ION, ... | | Authors: | Yamanaka, Y, Hashimoto, K, Noguchi, K, Yohda, M, Odaka, M. | | Deposit date: | 2014-12-10 | | Release date: | 2016-01-27 | | Method: | X-RAY DIFFRACTION (1.24 Å) | | Cite: | Time-Resolved Crystallography of the Reaction Intermediate of Nitrile Hydratase: Revealing a Role for the Cysteinesulfenic Acid Ligand as a Catalytic Nucleophile.
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
