3SS9
 
 | | Crystal structure of holo D-serine dehydratase from Escherichia coli at 1.97 A resolution | | Descriptor: | D-serine dehydratase, POTASSIUM ION, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Urusova, D.V, Isupov, M.N, Antonyuk, S.V, Kachalova, G.S, Vagin, A.A, Lebedev, A.A, Bourenkov, G.P, Dauter, Z, Bartunik, H.D, Melik-Adamyan, W.R, Mueller, T.D, Schnackerz, K.D. | | Deposit date: | 2011-07-08 | | Release date: | 2012-01-18 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Crystal structure of D-serine dehydratase from Escherichia coli.
Biochim.Biophys.Acta, 1824, 2011
|
|
4YEM
 
 | | Carboplatin binding to HEWL in NaBr crystallisation conditions studied at an X-ray wavelength of 0.9163A - new refinement | | Descriptor: | ACETATE ION, BROMIDE ION, CHLORIDE ION, ... | | Authors: | Shabalin, I.G, Dauter, Z, Jaskolski, M, Minor, W, Wlodawer, A. | | Deposit date: | 2015-02-24 | | Release date: | 2015-03-04 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Crystallography and chemistry should always go together: a cautionary tale of protein complexes with cisplatin and carboplatin.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
1HHF
 
 | | Decaplanin second P6122-Form | | Descriptor: | 4-epi-vancosamine, CHLORIDE ION, DECAPLANIN, ... | | Authors: | Lehmann, C, Vertessy, L, Sheldrick, G.M, Dauter, Z, Dauter, M. | | Deposit date: | 2000-12-22 | | Release date: | 2005-07-11 | | Last modified: | 2025-04-09 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Structures of Four Crystal Forms of Decaplanin
Helv.Chim.Acta, 86, 2003
|
|
4YEN
 
 | | Room temperature X-ray diffraction studies of cisplatin binding to HEWL in DMSO media after 14 months of crystal storage - new refinement | | Descriptor: | CHLORIDE ION, DIMETHYL SULFOXIDE, Lysozyme C, ... | | Authors: | Shabalin, I.G, Dauter, Z, Jaskolski, M, Minor, W, Wlodawer, A. | | Deposit date: | 2015-02-24 | | Release date: | 2015-03-04 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystallography and chemistry should always go together: a cautionary tale of protein complexes with cisplatin and carboplatin.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
1GQB
 
 | | HUMAN MIR-RECEPTOR, REPEAT 11 | | Descriptor: | BROMIDE ION, CATION-INDEPENDENT MANNOSE-6-PHOSPHATE RECEPTOR | | Authors: | Von Buelow, R, Dauter, M, Dauter, Z, Rajashankar, K.R, Grimme, S, Schmidt, B, Von Figura, K, Uson, I. | | Deposit date: | 2001-11-22 | | Release date: | 2002-12-05 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Locating the Anomalous Scatterer Substructures in Halide and Sulfur Phasing
Acta Crystallogr.,Sect.D, 59, 2003
|
|
4YEO
 
 | | Triclinic HEWL co-crystallised with cisplatin, studied at a data collection temperature of 150K - new refinement | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, Cisplatin, ... | | Authors: | Shabalin, I.G, Dauter, Z, Jaskolski, M, Minor, W, Wlodawer, A. | | Deposit date: | 2015-02-24 | | Release date: | 2015-03-04 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (0.98 Å) | | Cite: | Crystallography and chemistry should always go together: a cautionary tale of protein complexes with cisplatin and carboplatin.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
1HH3
 
 | | Decaplanin first P21-Form | | Descriptor: | 4-epi-vancosamine, DECAPLANIN, GLYCEROL, ... | | Authors: | Lehmann, C, Vertessy, L, Sheldrick, G.M, Dauter, Z, Dauter, M. | | Deposit date: | 2000-12-19 | | Release date: | 2005-07-11 | | Last modified: | 2025-04-09 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Structures of Four Crystal Forms of Decaplanin
Helv.Chim.Acta, 86, 2003
|
|
1HHC
 
 | | Crystal structure of Decaplanin - space group P21, second form | | Descriptor: | 4-epi-vancosamine, CITRIC ACID, DECAPLANIN, ... | | Authors: | Lehmann, C, Vertessy, L, Sheldrick, G.M, Dauter, Z, Dauter, M. | | Deposit date: | 2000-12-22 | | Release date: | 2005-07-11 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.13 Å) | | Cite: | Structures of Four Crystal Forms of Decaplanin
Helv.Chim.Acta, 86, 2003
|
|
2FBA
 
 | | Glucoamylase from Saccharomycopsis fibuligera at atomic resolution | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Glucoamylase GLU1 | | Authors: | Sevcik, J, Hostinova, E, Solovicova, A, Gasperik, J, Dauter, Z, Wilson, K.S. | | Deposit date: | 2005-12-09 | | Release date: | 2006-05-23 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Structure of the complex of a yeast glucoamylase with acarbose reveals the presence of a raw starch binding site on the catalytic domain.
Febs J., 273, 2006
|
|
4YDX
 
 | | Crystal structure of cisplatin bound to a human copper chaperone (monomer) - new refinement | | Descriptor: | 3,3',3''-phosphanetriyltripropanoic acid, Copper transport protein ATOX1, PLATINUM (II) ION, ... | | Authors: | Shabalin, I.G, Boal, A.K, Dauter, Z, Jaskolski, M, Minor, W, Rosenzweig, A.C, Wlodawer, A. | | Deposit date: | 2015-02-23 | | Release date: | 2015-03-04 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.602 Å) | | Cite: | Crystallography and chemistry should always go together: a cautionary tale of protein complexes with cisplatin and carboplatin.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4YEA
 
 | | Crystal structure of cisplatin bound to a human copper chaperone (dimer) - new refinement | | Descriptor: | COPPER (II) ION, Copper transport protein ATOX1, SULFATE ION | | Authors: | Shabalin, I.G, Dauter, Z, Jaskolski, M, Minor, W, Wlodawer, A. | | Deposit date: | 2015-02-23 | | Release date: | 2015-03-18 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Crystallography and chemistry should always go together: a cautionary tale of protein complexes with cisplatin and carboplatin.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
5H8K
 
 | | Crystal structure of Medicago truncatula N-carbamoylputrescine amidohydrolase (MtCPA) C158S mutant | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Sekula, B, Ruszkowski, M, Malinska, M, Dauter, Z. | | Deposit date: | 2015-12-23 | | Release date: | 2016-04-20 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Structural Investigations of N-carbamoylputrescine Amidohydrolase from Medicago truncatula: Insights into the Ultimate Step of Putrescine Biosynthesis in Plants.
Front Plant Sci, 7, 2016
|
|
5H8I
 
 | | Crystal structure of Medicago truncatula N-carbamoylputrescine amidohydrolase (MtCPA) in complex with N-(dihydroxymethyl)putrescine | | Descriptor: | (4-azanylbutylamino)methanediol, 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Sekula, B, Ruszkowski, M, Malinska, M, Dauter, Z. | | Deposit date: | 2015-12-23 | | Release date: | 2016-04-20 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Structural Investigations of N-carbamoylputrescine Amidohydrolase from Medicago truncatula: Insights into the Ultimate Step of Putrescine Biosynthesis in Plants.
Front Plant Sci, 7, 2016
|
|
5H8J
 
 | | Crystal structure of Medicago truncatula N-carbamoylputrescine amidohydrolase (MtCPA) in complex with cadaverine | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Sekula, B, Ruszkowski, M, Malinska, M, Dauter, Z. | | Deposit date: | 2015-12-23 | | Release date: | 2016-04-20 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Structural Investigations of N-carbamoylputrescine Amidohydrolase from Medicago truncatula: Insights into the Ultimate Step of Putrescine Biosynthesis in Plants.
Front Plant Sci, 7, 2016
|
|
5H8L
 
 | | Crystal structure of Medicago truncatula N-carbamoylputrescine amidohydrolase (MtCPA) C158S mutant in complex with putrescine | | Descriptor: | 1,2-ETHANEDIOL, 1,4-DIAMINOBUTANE, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Sekula, B, Ruszkowski, M, Malinska, M, Dauter, Z. | | Deposit date: | 2015-12-23 | | Release date: | 2016-04-20 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Structural Investigations of N-carbamoylputrescine Amidohydrolase from Medicago truncatula: Insights into the Ultimate Step of Putrescine Biosynthesis in Plants.
Front Plant Sci, 7, 2016
|
|
2TEC
 
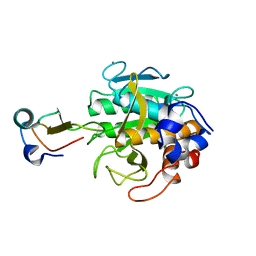 | | MOLECULAR DYNAMICS REFINEMENT OF A THERMITASE-EGLIN-C COMPLEX AT 1.98 ANGSTROMS RESOLUTION AND COMPARISON OF TWO CRYSTAL FORMS THAT DIFFER IN CALCIUM CONTENT | | Descriptor: | CALCIUM ION, EGLIN C, THERMITASE | | Authors: | Gros, P, Betzel, C, Dauter, Z, Wilson, K.S, Hol, W.G.J. | | Deposit date: | 1990-10-26 | | Release date: | 1992-01-15 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Molecular dynamics refinement of a thermitase-eglin-c complex at 1.98 A resolution and comparison of two crystal forms that differ in calcium content.
J.Mol.Biol., 210, 1989
|
|
2JHF
 
 | | Structural evidence for a ligand coordination switch in liver alcohol dehydrogenase | | Descriptor: | ALCOHOL DEHYDROGENASE E CHAIN, CADMIUM ION, DIMETHYL SULFOXIDE, ... | | Authors: | Meijers, R, Adolph, H.W, Dauter, Z, Wilson, K.S, Lamzin, V.S, Cedergren-Zeppezauer, E.S. | | Deposit date: | 2007-02-22 | | Release date: | 2007-04-24 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Structural Evidence for a Ligand Coordination Switch in Liver Alcohol Dehydrogenase
Biochemistry, 46, 2007
|
|
2JHG
 
 | | Structural evidence for a ligand coordination switch in liver alcohol dehydrogenase | | Descriptor: | 2-METHYLPROPANAMIDE, ALCOHOL DEHYDROGENASE E CHAIN, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Meijers, R, Adolph, H.W, Dauter, Z, Wilson, K.S, Lamzin, V.S, Cedergren-Zeppezauer, E.S. | | Deposit date: | 2007-02-22 | | Release date: | 2007-04-24 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Structural Evidence for a Ligand Coordination Switch in Liver Alcohol Dehydrogenase
Biochemistry, 46, 2007
|
|
4LZT
 
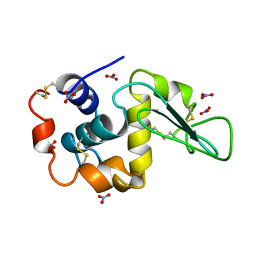 | | ATOMIC RESOLUTION REFINEMENT OF TRICLINIC HEW LYSOZYME AT 295K | | Descriptor: | LYSOZYME, NITRATE ION | | Authors: | Walsh, M.A, Schneider, T, Sieker, L.C, Dauter, Z, Lamzin, V, Wilson, K.S. | | Deposit date: | 1997-03-31 | | Release date: | 1998-04-01 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (0.95 Å) | | Cite: | Refinement of triclinic hen egg-white lysozyme at atomic resolution.
Acta Crystallogr.,Sect.D, 54, 1998
|
|
1E6F
 
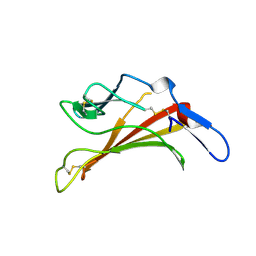 | | Human MIR-receptor, repeat 11 | | Descriptor: | CATION-INDEPENDENT MANNOSE-6-PHOSPHATE RECEPTOR | | Authors: | Von Buelow, R, Rajashankar, K.R, Dauter, M, Dauter, Z, Grimme, S, Schmidt, B, Von Figura, K, Uson, I. | | Deposit date: | 2000-08-15 | | Release date: | 2001-08-09 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Locating the Anomalous Scatterer Substructures in Halide and Sulfur Phasing
Acta Crystallogr.,Sect.D, 59, 2003
|
|
4N3E
 
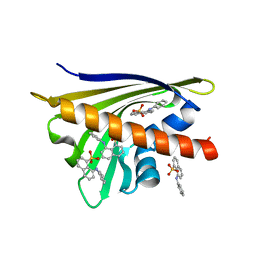 | | Crystal structure of Hyp-1, a St John's wort PR-10 protein, in complex with 8-anilino-1-naphthalene sulfonate (ANS) | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, 8-ANILINO-1-NAPHTHALENE SULFONATE, Phenolic oxidative coupling protein, ... | | Authors: | Sliwiak, J, Dauter, Z, Mccoy, A.J, Read, R.J, Jaskolski, M. | | Deposit date: | 2013-10-07 | | Release date: | 2014-02-26 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | Likelihood-based molecular-replacement solution for a highly pathological crystal with tetartohedral twinning and sevenfold translational noncrystallographic symmetry.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
2NAC
 
 | | HIGH RESOLUTION STRUCTURES OF HOLO AND APO FORMATE DEHYDROGENASE | | Descriptor: | NAD-DEPENDENT FORMATE DEHYDROGENASE, SULFATE ION | | Authors: | Lamzin, V.S, Dauter, Z, Popov, V.O, Harutyunyan, E.H, Wilson, K.S. | | Deposit date: | 1994-07-06 | | Release date: | 1995-01-26 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | High resolution structures of holo and apo formate dehydrogenase.
J.Mol.Biol., 236, 1994
|
|
2NAD
 
 | | HIGH RESOLUTION STRUCTURES OF HOLO AND APO FORMATE DEHYDROGENASE | | Descriptor: | AZIDE ION, NAD-DEPENDENT FORMATE DEHYDROGENASE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Lamzin, V.S, Dauter, Z, Popov, V.O, Harutyunyan, E.H, Wilson, K.S. | | Deposit date: | 1994-07-06 | | Release date: | 1995-01-26 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | High resolution structures of holo and apo formate dehydrogenase.
J.Mol.Biol., 236, 1994
|
|
6VCW
 
 | | Crystal structure of Medicago truncatula S-adenosylmethionine Synthase 3A (MtMAT3A) | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Sekula, B, Ruszkowski, M, Dauter, Z. | | Deposit date: | 2019-12-23 | | Release date: | 2020-02-26 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | S-adenosylmethionine synthases in plants: Structural characterization of type I and II isoenzymes from Arabidopsis thaliana and Medicago truncatula.
Int.J.Biol.Macromol., 151, 2020
|
|
6VD2
 
 | | Crystal structure of Arabidopsis thaliana S-adenosylmethionine Synthase 2 (AtMAT2) in complex with S-adenosylmethionine | | Descriptor: | 3,6,9,12,15,18,21-HEPTAOXATRICOSANE-1,23-DIOL, 3[N-MORPHOLINO]PROPANE SULFONIC ACID, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Sekula, B, Ruszkowski, M, Dauter, Z. | | Deposit date: | 2019-12-23 | | Release date: | 2020-02-26 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | S-adenosylmethionine synthases in plants: Structural characterization of type I and II isoenzymes from Arabidopsis thaliana and Medicago truncatula.
Int.J.Biol.Macromol., 151, 2020
|
|
