7O5B
 
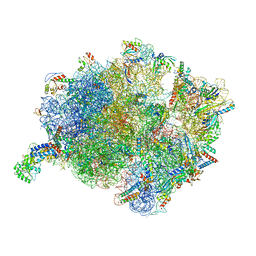 | | Cryo-EM structure of a Bacillus subtilis MifM-stalled ribosome-nascent chain complex with (p)ppGpp-SRP bound | | Descriptor: | 16S rRNA (1533-MER), 23S rRNA (2887-MER), 30S ribosomal protein S10, ... | | Authors: | Kratzat, H, Czech, L, Berninghausen, O, Bange, G, Beckmann, R. | | Deposit date: | 2021-04-08 | | Release date: | 2022-02-02 | | Last modified: | 2022-03-09 | | Method: | ELECTRON MICROSCOPY (3.33 Å) | | Cite: | Inhibition of SRP-dependent protein secretion by the bacterial alarmone (p)ppGpp.
Nat Commun, 13, 2022
|
|
5DEC
 
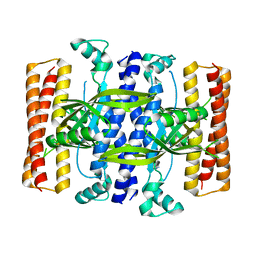 | |
7OD9
 
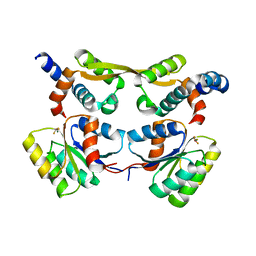 | | Crystal structure of activated CheY fused to the C-terminal domain of CheF | | Descriptor: | BERYLLIUM TRIFLUORIDE ION, C-terminal domain of CheF from Methanococcus maripaludis, MAGNESIUM ION, ... | | Authors: | Altegoer, F, Weiland, P, Bange, G. | | Deposit date: | 2021-04-29 | | Release date: | 2022-04-27 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural insights into the mechanism of archaellar rotational switching.
Nat Commun, 13, 2022
|
|
7P1W
 
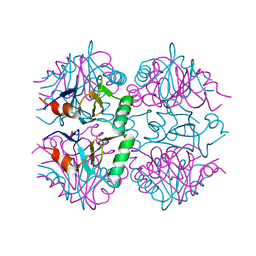 | |
5AFF
 
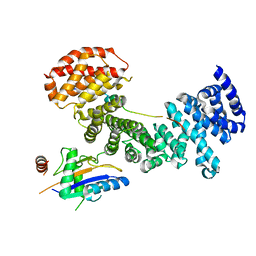 | | Symportin 1 chaperones 5S RNP assembly during ribosome biogenesis by occupying an essential rRNA binding site | | Descriptor: | RIBOSOMAL PROTEIN L11, RIBOSOMAL PROTEIN L5, SYMPORTIN 1 | | Authors: | Calvino, F.R, Kharde, S, Wild, K, Bange, G, Sinning, I. | | Deposit date: | 2015-01-21 | | Release date: | 2015-04-15 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.398 Å) | | Cite: | Symportin 1 Chaperones 5S Rnp Assembly During Ribosome Biogenesis by Occupying an Essential Rrna-Binding Site.
Nat.Commun., 6, 2015
|
|
5DED
 
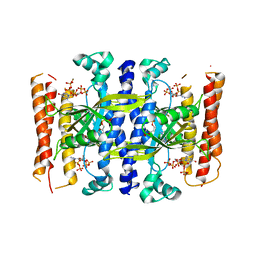 | | Crystal structure of the small alarmone synthethase 1 from Bacillus subtilis bound to its product pppGpp | | Descriptor: | GTP pyrophosphokinase YjbM, MAGNESIUM ION, guanosine 5'-(tetrahydrogen triphosphate) 3'-(trihydrogen diphosphate) | | Authors: | Steinchen, W, Schuhmacher, J.S, Altegoer, F, Bange, G. | | Deposit date: | 2015-08-25 | | Release date: | 2015-10-28 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.942 Å) | | Cite: | Catalytic mechanism and allosteric regulation of an oligomeric (p)ppGpp synthetase by an alarmone.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
7O9F
 
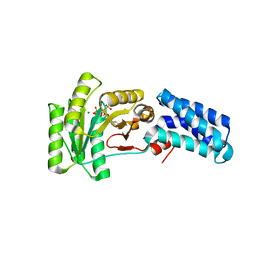 | | Bacillus subtilis Ffh in complex with ppGpp | | Descriptor: | GUANOSINE-5',3'-TETRAPHOSPHATE, MAGNESIUM ION, Signal recognition particle protein | | Authors: | Czech, L, Mais, C.-N, Bange, G. | | Deposit date: | 2021-04-16 | | Release date: | 2022-02-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Inhibition of SRP-dependent protein secretion by the bacterial alarmone (p)ppGpp.
Nat Commun, 13, 2022
|
|
7O9I
 
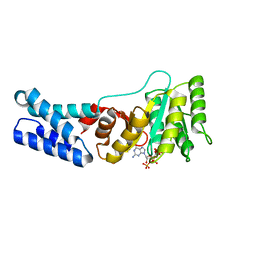 | | Escherichia coli Ffh in complex with pppGpp | | Descriptor: | Signal recognition particle protein, guanosine 5'-(tetrahydrogen triphosphate) 3'-(trihydrogen diphosphate) | | Authors: | Czech, L, Mais, C.-N, Bange, G. | | Deposit date: | 2021-04-16 | | Release date: | 2022-02-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Inhibition of SRP-dependent protein secretion by the bacterial alarmone (p)ppGpp.
Nat Commun, 13, 2022
|
|
7O9G
 
 | | Escherichia coli Ffh in complex with ppGpp | | Descriptor: | GUANOSINE-5',3'-TETRAPHOSPHATE, MAGNESIUM ION, Signal recognition particle protein | | Authors: | Czech, L, Mais, C.-N, Bange, G. | | Deposit date: | 2021-04-16 | | Release date: | 2022-02-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Inhibition of SRP-dependent protein secretion by the bacterial alarmone (p)ppGpp.
Nat Commun, 13, 2022
|
|
7O9H
 
 | | Escherichia coli FtsY in complex with pppGpp | | Descriptor: | Signal recognition particle receptor FtsY, guanosine 5'-(tetrahydrogen triphosphate) 3'-(trihydrogen diphosphate) | | Authors: | Czech, L, Mais, C.-N, Bange, G. | | Deposit date: | 2021-04-16 | | Release date: | 2022-02-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Inhibition of SRP-dependent protein secretion by the bacterial alarmone (p)ppGpp.
Nat Commun, 13, 2022
|
|
7OVP
 
 | |
5DMB
 
 | |
5DMD
 
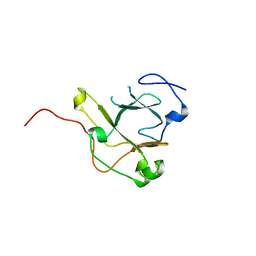 | | Crystal structure of the flagellar assembly factor FliW | | Descriptor: | Flagellar assembly factor FliW | | Authors: | Altegoer, F, Bange, G. | | Deposit date: | 2015-09-08 | | Release date: | 2016-09-07 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structural basis for the CsrA-dependent modulation of translation initiation by an ancient regulatory protein.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
7BM2
 
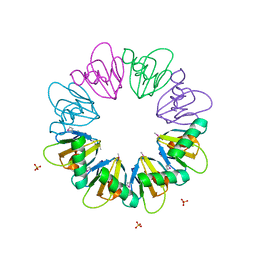 | |
7BME
 
 | |
6EKH
 
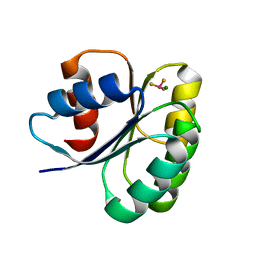 | |
6EKG
 
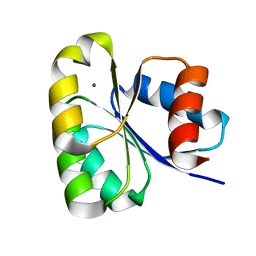 | |
6EN7
 
 | |
6FPK
 
 | | Co-translational folding intermediate dictates membrane targeting of the signal recognition particle (SRP)- receptor | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Signal recognition particle receptor FtsY | | Authors: | Karniel, A, Mrusek, D, Steinchen, W, Dym, O, Bange, G, Bibi, E. | | Deposit date: | 2018-02-11 | | Release date: | 2018-05-09 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Co-translational Folding Intermediate Dictates Membrane Targeting of the Signal Recognition Particle Receptor.
J. Mol. Biol., 430, 2018
|
|
6FPR
 
 | | Co-translational folding intermediate dictates membrane targeting of the signal recognition particle (SRP)- receptor | | Descriptor: | Signal recognition particle receptor FtsY | | Authors: | Karniel, A, Mrusek, D, Steinchen, W, Dym, O, Bange, G, Bibi, E. | | Deposit date: | 2018-02-12 | | Release date: | 2018-05-09 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Co-translational Folding Intermediate Dictates Membrane Targeting of the Signal Recognition Particle Receptor.
J. Mol. Biol., 430, 2018
|
|
6FPG
 
 | | Structure of the Ustilago maydis chorismate mutase 1 in complex with a Zea mays kiwellin | | Descriptor: | CITRIC ACID, Chromosome 16, whole genome shotgun sequence, ... | | Authors: | Altegoer, F, Steinchen, W, Bange, G. | | Deposit date: | 2018-02-09 | | Release date: | 2019-01-16 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A kiwellin disarms the metabolic activity of a secreted fungal virulence factor.
Nature, 565, 2019
|
|
6G0Z
 
 | | Crystal structure of GDP bound RbgA from S. aureus | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, Ribosome biogenesis GTPase A | | Authors: | Pausch, P, Bange, G. | | Deposit date: | 2018-03-20 | | Release date: | 2018-11-07 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structural basis for (p)ppGpp-mediated inhibition of the GTPase RbgA.
J. Biol. Chem., 293, 2018
|
|
6G15
 
 | | Crystal structure of pppGpp bound RbgA from S. aureus | | Descriptor: | Ribosome biogenesis GTPase A, guanosine 5'-(tetrahydrogen triphosphate) 3'-(trihydrogen diphosphate) | | Authors: | Pausch, P, Bange, G. | | Deposit date: | 2018-03-20 | | Release date: | 2018-11-07 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural basis for (p)ppGpp-mediated inhibition of the GTPase RbgA.
J. Biol. Chem., 293, 2018
|
|
6G12
 
 | | Crystal structure of GMPPNP bound RbgA from S. aureus | | Descriptor: | PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER, Ribosome biogenesis GTPase A | | Authors: | Pausch, P, Bange, G. | | Deposit date: | 2018-03-20 | | Release date: | 2018-11-07 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.929 Å) | | Cite: | Structural basis for (p)ppGpp-mediated inhibition of the GTPase RbgA.
J. Biol. Chem., 293, 2018
|
|
6FPF
 
 | |
