4EMR
 
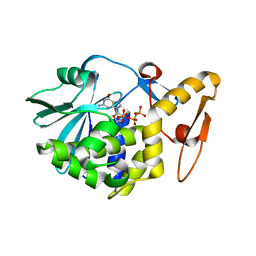 | | Crystal Structure determination of type1 ribosome inactivating protein complexed with 7-methylguanosine-triphosphate at 1.75A | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 7-METHYL-GUANOSINE-5'-TRIPHOSPHATE, rRNA N-glycosidase | | Authors: | Kumar, M, Kushwaha, G.S, Sinha, M, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2012-04-12 | | Release date: | 2012-05-02 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | First structural evidence of sequestration of mRNA cap structures by type 1 ribosome inactivating protein from Momordica balsamina.
Proteins, 81, 2013
|
|
3UMQ
 
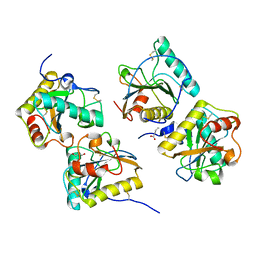 | | Crystal structure of peptidoglycan recognition protein-S complexed with butyric acid at 2.2 A resolution | | Descriptor: | GLYCEROL, Peptidoglycan recognition protein 1, butanoic acid | | Authors: | Pandey, N, Sharma, P, Sinha, M, Bhushan, A, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2011-11-14 | | Release date: | 2012-07-04 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis of the binding of fatty acids to peptidoglycan recognition protein, PGRP-S through second binding site
Arch.Biochem.Biophys., 529, 2013
|
|
3UIL
 
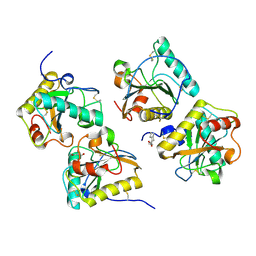 | | Crystal Structure of the complex of PGRP-S with lauric acid at 2.2 A resolution | | Descriptor: | GLYCEROL, LAURIC ACID, Peptidoglycan recognition protein 1 | | Authors: | Dube, D, Sharma, P, Sinha, M, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2011-11-05 | | Release date: | 2012-07-11 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis of the binding of fatty acids to peptidoglycan recognition protein, PGRP-S through second binding site
Arch.Biochem.Biophys., 529, 2013
|
|
3T2V
 
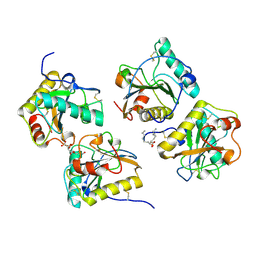 | | Crystal structure of the complex of peptidoglycan recognition protein-short (CPGRP-S) with mycolic acid at 2.5 A resolution | | Descriptor: | (2S,3R)-2-hexyl-3-hydroxynonanoic acid, GLYCEROL, L(+)-TARTARIC ACID, ... | | Authors: | Sharma, P, Dube, D, Sinha, M, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2011-07-23 | | Release date: | 2011-08-10 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Structural basis of the binding of fatty acids to peptidoglycan recognition protein, PGRP-S through second binding site
Arch.Biochem.Biophys., 529, 2013
|
|
9FAB
 
 | | Additional cryo-EM structure of cardiac amyloid AL59 - bent polymorph | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Monoclonal immunoglobulin light chains (LC) | | Authors: | Schulte, T, Speranzini, V, Chaves-Sanjuan, A, Milazzo, M, Ricagno, S. | | Deposit date: | 2024-05-10 | | Release date: | 2024-08-14 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Additional cryo-EM structure of cardiac amyloid AL59 - bent polymorph
To be published
|
|
9FAC
 
 | | Additional cryo-EM structure of cardiac amyloid AL59 - mixed polymorph | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Monoclonal immunoglobulin light chains (LC) | | Authors: | Schulte, T, Speranzini, V, Chaves-Sanjuan, A, Milazzo, M, Ricagno, S. | | Deposit date: | 2024-05-10 | | Release date: | 2024-08-14 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Helical superstructures between amyloid and collagen in cardiac fibrils from a patient with AL amyloidosis.
Nat Commun, 15, 2024
|
|
9FAA
 
 | | Cryo-EM structure of cardiac collagen-associated amyloid AL59 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Monoclonal immunoglobulin light chains (LC) | | Authors: | Schulte, T, Speranzini, V, Chaves-Sanjuan, A, Milazzo, M, Ricagno, S. | | Deposit date: | 2024-05-10 | | Release date: | 2024-07-17 | | Last modified: | 2024-08-14 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Helical superstructures between amyloid and collagen in cardiac fibrils from a patient with AL amyloidosis.
Nat Commun, 15, 2024
|
|
