2AXZ
 
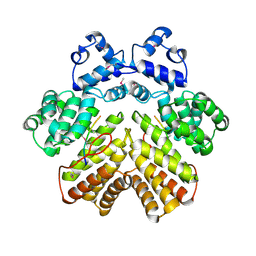 | | Crystal structure of PrgX/cCF10 complex | | Descriptor: | LVTLVFV peptide, PrgX, TPPKEVT(MSE) peptide | | Authors: | Shi, K, Brown, C.K, Gu, Z.Y, Kozlowicz, B.K, Dunny, G.M, Ohlendorf, D.H, Earhart, C.A. | | Deposit date: | 2005-09-06 | | Release date: | 2005-12-06 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure of peptide sex pheromone receptor PrgX and PrgX/pheromone complexes and regulation of conjugation in Enterococcus faecalis.
Proc.Natl.Acad.Sci.Usa, 102, 2005
|
|
2AXV
 
 | | Structure of PrgX Y153C mutant | | Descriptor: | PrgX | | Authors: | Shi, K, Brown, C.K, Gu, Z.Y, Kozlowicz, B.K, Dunny, G.M, Ohlendorf, D.H, Earhart, C.A. | | Deposit date: | 2005-09-06 | | Release date: | 2005-12-06 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure of peptide sex pheromone receptor PrgX and PrgX/pheromone complexes and regulation of conjugation in Enterococcus faecalis.
Proc.Natl.Acad.Sci.Usa, 102, 2005
|
|
2AW6
 
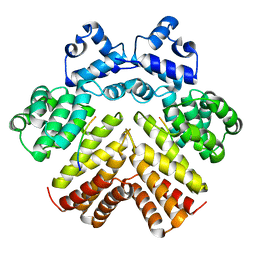 | | Structure of a bacterial peptide pheromone/receptor complex and its mechanism of gene regulation | | Descriptor: | PrgX, peptide | | Authors: | Shi, K, Brown, C.K, Gu, Z.Y, Kozlowicz, B.K, Dunny, G.M, Ohlendorf, D.H, Earhart, C.A. | | Deposit date: | 2005-08-31 | | Release date: | 2005-12-06 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure of peptide sex pheromone receptor PrgX and PrgX/pheromone complexes and regulation of conjugation in Enterococcus faecalis.
Proc.Natl.Acad.Sci.Usa, 102, 2005
|
|
1TS2
 
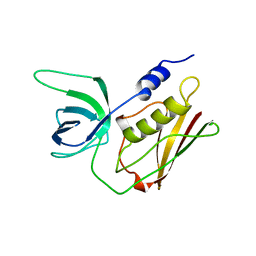 | | T128A MUTANT OF TOXIC SHOCK SYNDROME TOXIN-1 FROM S. AUREUS | | Descriptor: | TOXIC SHOCK SYNDROME TOXIN-1 | | Authors: | Earhart, C.A, Mitchell, D.T, Murray, D.L, Pinheiro, D.M, Matsumura, M, Schlievert, P.M, Ohlendorf, D.H. | | Deposit date: | 1997-10-09 | | Release date: | 1998-12-16 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structures of five mutants of toxic shock syndrome toxin-1 with reduced biological activity.
Biochemistry, 37, 1998
|
|
1TS3
 
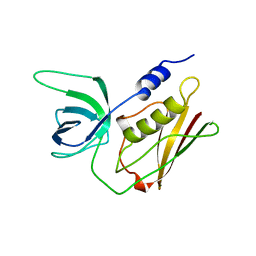 | | H135A MUTANT OF TOXIC SHOCK SYNDROME TOXIN-1 FROM S. AUREUS | | Descriptor: | TOXIC SHOCK SYNDROME TOXIN-1 | | Authors: | Earhart, C.A, Mitchell, D.T, Murray, D.L, Pinheiro, D.M, Matsumura, M, Schlievert, P.M, Ohlendorf, D.H. | | Deposit date: | 1997-10-10 | | Release date: | 1998-12-16 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structures of five mutants of toxic shock syndrome toxin-1 with reduced biological activity.
Biochemistry, 37, 1998
|
|
1RSM
 
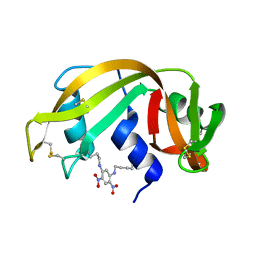 | | THE 2-ANGSTROMS RESOLUTION STRUCTURE OF A THERMOSTABLE RIBONUCLEASE A CHEMICALLY CROSS-LINKED BETWEEN LYSINE RESIDUES 7 AND 41 | | Descriptor: | DINITROPHENYLENE, RIBONUCLEASE A | | Authors: | Weber, P.C, Sheriff, S, Ohlendorf, D.H, Finzel, B.C, Salemme, F.R. | | Deposit date: | 1985-08-27 | | Release date: | 1986-01-21 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The 2-A resolution structure of a thermostable ribonuclease A chemically cross-linked between lysine residues 7 and 41.
Proc.Natl.Acad.Sci.USA, 82, 1985
|
|
1EXF
 
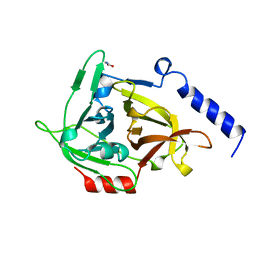 | | EXFOLIATIVE TOXIN A | | Descriptor: | EXFOLIATVE TOXIN A, GLYCINE | | Authors: | Vath, G.M, Earhart, C.A, Rago, J.V, Kim, M.H, Bohach, G.A, Schlievert, P.M, Ohlendorf, D.H. | | Deposit date: | 1996-10-22 | | Release date: | 1998-02-25 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The structure of the superantigen exfoliative toxin A suggests a novel regulation as a serine protease.
Biochemistry, 36, 1997
|
|
1F1R
 
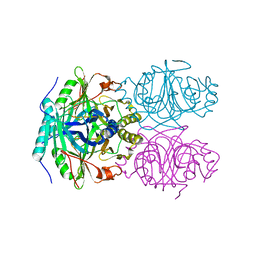 | | CRYSTAL STRUCTURE OF HOMOPROTOCATECHUATE 2,3-DIOXYGENASE FROM ARTHROBACTER GLOBIFORMIS (NATIVE, NON-CRYO) | | Descriptor: | HOMOPROTOCATECHUATE 2,3-DIOXYGENASE, MANGANESE (II) ION | | Authors: | Vetting, M.W, Lipscomb, J.D, Wackett, L.P, Que Jr, L, Ohlendorf, D.H. | | Deposit date: | 2000-05-19 | | Release date: | 2003-06-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystallographic comparison of manganese- and iron-dependent homoprotocatechuate 2,3-dioxygenases.
J.Bacteriol., 186, 2004
|
|
1LRP
 
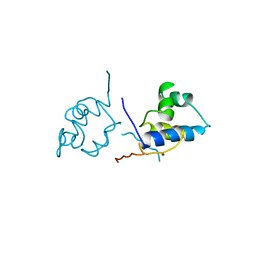 | |
8BTY
 
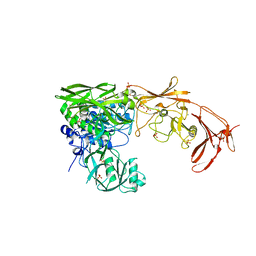 | | Structure of the active form of ScpB, the C5a-peptidase from Streptococcus agalactiae. | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, C5a peptidase, CALCIUM ION, ... | | Authors: | Kagawa, T.F, Cooney, J.C, Miclot, T, Cullen, R. | | Deposit date: | 2022-11-30 | | Release date: | 2023-11-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The 1.7 angstrom crystal structure of the C5a peptidase from Streptococcus agalactiae (ScpB) reveals an active site competent for catalysis.
Proteins, 92, 2024
|
|
3LMX
 
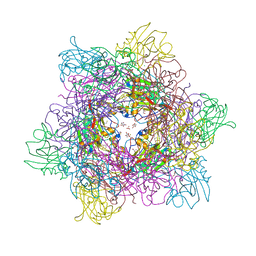 | | Tyrosine 447 of Protocatechuate 34,-Dioxygenase Controls Efficient Progress Through Catalysis | | Descriptor: | 3,4-DIHYDROXYBENZOIC ACID, BETA-MERCAPTOETHANOL, CHLORIDE ION, ... | | Authors: | Purpero, V.M, Lipscomb, J.D, Shi, K. | | Deposit date: | 2010-02-01 | | Release date: | 2011-02-16 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Tyrosine 447 of Protocatechuate 34,-Dioxygenase Controls Efficient Progress Through Catalysis
To be Published
|
|
3LKT
 
 | |
3LXV
 
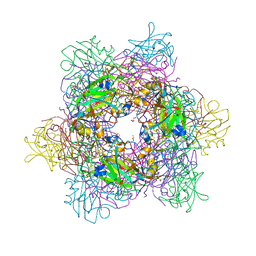 | | Tyrosine 447 of Protocatechuate 3,4-Dioxygenase Controls Efficient Progress Through Catalysis | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 4-NITROCATECHOL, BETA-MERCAPTOETHANOL, ... | | Authors: | Purpero, V.M, Lipscomb, J.D. | | Deposit date: | 2010-02-25 | | Release date: | 2011-03-09 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Tyrosine 447 of Protocatechuate 3,4-Dioxygenase Controls Efficient Progress Through Catalysis
To be Published, 2010
|
|
1ESF
 
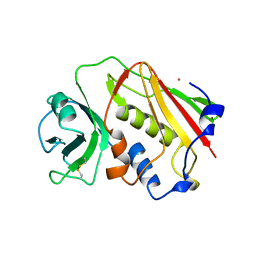 | | STAPHYLOCOCCAL ENTEROTOXIN A | | Descriptor: | CADMIUM ION, STAPHYLOCOCCAL ENTEROTOXIN A | | Authors: | Schad, E.M, Svensson, L.A. | | Deposit date: | 1995-05-25 | | Release date: | 1996-07-11 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of the superantigen staphylococcal enterotoxin type A.
EMBO J., 14, 1995
|
|
1YLC
 
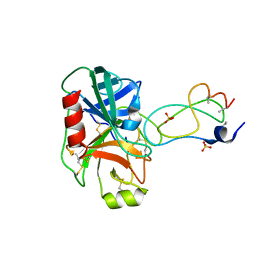 | | Trypsin/BPTI complex mutant | | Descriptor: | CALCIUM ION, Pancreatic trypsin inhibitor, SULFATE ION, ... | | Authors: | Brown, C.K, Ohlendorf, D.H. | | Deposit date: | 2005-01-19 | | Release date: | 2006-04-25 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Partially folded bovine pancreatic trypsin inhibitor analogues attain fully native structures when co-crystallized with S195A rat trypsin
J.Mol.Biol., 375, 2008
|
|
1YKT
 
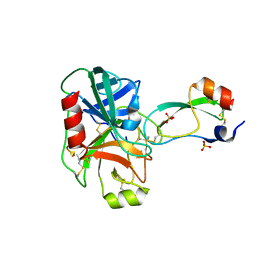 | | Trypsin/Bpti complex mutant | | Descriptor: | CALCIUM ION, Pancreatic trypsin inhibitor, SULFATE ION, ... | | Authors: | Brown, C.K, Ohlendorf, D.H. | | Deposit date: | 2005-01-18 | | Release date: | 2006-04-25 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Partially folded bovine pancreatic trypsin inhibitor analogues attain fully native structures when co-crystallized with S195A rat trypsin
J.Mol.Biol., 375, 2008
|
|
1YLD
 
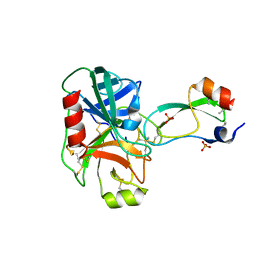 | | Trypsin/BPTI complex mutant | | Descriptor: | CALCIUM ION, Pancreatic trypsin inhibitor, SULFATE ION, ... | | Authors: | Brown, C.K, Ohlendorf, D.H. | | Deposit date: | 2005-01-19 | | Release date: | 2006-04-25 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Partially folded bovine pancreatic trypsin inhibitor analogues attain fully native structures when co-crystallized with S195A rat trypsin
J.Mol.Biol., 375, 2008
|
|
6CRO
 
 | |
1STP
 
 | |
4CRO
 
 | | PROTEIN-DNA CONFORMATIONAL CHANGES IN THE CRYSTAL STRUCTURE OF A LAMBDA CRO-OPERATOR COMPLEX | | Descriptor: | DNA (5'-D(*TP*AP*TP*CP*AP*CP*CP*GP*CP*GP*GP*GP*TP*GP*AP*TP*A)-3'), PROTEIN (LAMBDA CRO) | | Authors: | Brennan, R.G, Roderick, S.L, Takeda, Y, Matthews, B.W. | | Deposit date: | 1992-01-15 | | Release date: | 1992-01-15 | | Last modified: | 2022-11-23 | | Method: | X-RAY DIFFRACTION (3.9 Å) | | Cite: | Protein-DNA conformational changes in the crystal structure of a lambda Cro-operator complex.
Proc.Natl.Acad.Sci.USA, 87, 1990
|
|
3ORC
 
 | |
1JCK
 
 | |
1LMB
 
 | | REFINED 1.8 ANGSTROM CRYSTAL STRUCTURE OF THE LAMBDA REPRESSOR-OPERATOR COMPLEX | | Descriptor: | DNA (5'-D(*AP*AP*TP*AP*CP*CP*AP*CP*TP*GP*GP*CP*GP*GP*TP*GP*A P*TP*AP*T)-3'), DNA (5'-D(*TP*AP*TP*AP*TP*CP*AP*CP*CP*GP*CP*CP*AP*GP*TP*GP*G P*TP*AP*T)-3'), PROTEIN (LAMBDA REPRESSOR) | | Authors: | Beamer, L.J, Pabo, C.O. | | Deposit date: | 1991-11-05 | | Release date: | 1991-11-05 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Refined 1.8 A crystal structure of the lambda repressor-operator complex.
J.Mol.Biol., 227, 1992
|
|
1QWX
 
 | | Crystal Structure of a Staphylococcal Inhibitor/Chaperone | | Descriptor: | cysteine protease | | Authors: | Brown, C.K, Gu, Z.-Y, Nickerson, N, McGavin, M.J, Ohlendorf, D.H, Earhart, C.A. | | Deposit date: | 2003-09-03 | | Release date: | 2004-02-10 | | Last modified: | 2014-03-12 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: |
|
|
1PTS
 
 | |
