6FPT
 
 | | Crystal structure of Danio rerio Lin41 filamin-NHL domains | | Descriptor: | E3 ubiquitin-protein ligase TRIM71 | | Authors: | Kumari, P, Aeschimann, F, Gaidatzis, D, Keusch, J.J, Ghosh, P, Neagu, A, Pachulska-Wieczorek, K, Bujnicki, J.M, Gut, H, Grosshans, H, Ciosk, R. | | Deposit date: | 2018-02-12 | | Release date: | 2018-05-09 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Evolutionary plasticity of the NHL domain underlies distinct solutions to RNA recognition.
Nat Commun, 9, 2018
|
|
6FQL
 
 | | Crystal structure of Danio rerio Lin41 filamin-NHL domains in complex with mab-10 3'UTR 13mer RNA | | Descriptor: | CHLORIDE ION, E3 ubiquitin-protein ligase TRIM71, RNA (5'-R(*UP*GP*CP*AP*UP*UP*UP*AP*AP*UP*GP*CP*A)-3') | | Authors: | Kumari, P, Aeschimann, F, Gaidatzis, D, Keusch, J.J, Ghosh, P, Neagu, A, Pachulska-Wieczorek, K, Bujnicki, J.M, Gut, H, Grosshans, H, Ciosk, R. | | Deposit date: | 2018-02-14 | | Release date: | 2018-05-09 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.349 Å) | | Cite: | Evolutionary plasticity of the NHL domain underlies distinct solutions to RNA recognition.
Nat Commun, 9, 2018
|
|
8EIT
 
 | | Structure of FFAR1-Gq complex bound to DHA | | Descriptor: | A modified Guanine nucleotide-binding protein G(q) subunit alpha, DOCOSA-4,7,10,13,16,19-HEXAENOIC ACID, Free fatty acid receptor 1, ... | | Authors: | Kumari, P, Inoue, A, Chapman, K, Lian, P, Rosenbaum, D.M. | | Deposit date: | 2022-09-15 | | Release date: | 2023-05-24 | | Last modified: | 2023-05-31 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Molecular mechanism of fatty acid activation of FFAR1.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8EJK
 
 | | Structure of FFAR1-Gq complex bound to TAK-875 in a lipid nanodisc | | Descriptor: | A modified Guanine nucleotide-binding protein G(q) subunit alpha, Free fatty acid receptor 1, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Kumari, P, Inoue, A, Chapman, K, Lian, P, Rosenbaum, D.M. | | Deposit date: | 2022-09-17 | | Release date: | 2023-05-24 | | Last modified: | 2023-05-31 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Molecular mechanism of fatty acid activation of FFAR1.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8EJC
 
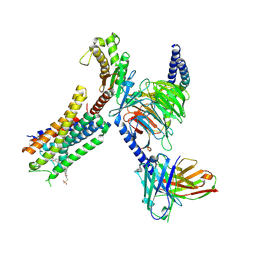 | | Structure of FFAR1-Gq complex bound to TAK-875 | | Descriptor: | A modified Guanine nucleotide-binding protein G(q) subunit alpha, Free fatty acid receptor 1, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Kumari, P, Inoue, A, Chapman, K, Lian, P, Rosenbaum, D.M. | | Deposit date: | 2022-09-16 | | Release date: | 2023-05-24 | | Last modified: | 2023-05-31 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Molecular mechanism of fatty acid activation of FFAR1.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
6LE1
 
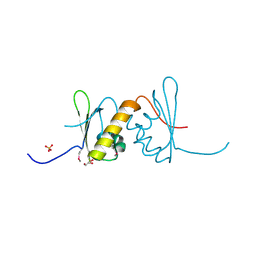 | | Structure of RRM2 domain of DND1 protein | | Descriptor: | Dead end protein homolog 1, GLYCEROL, SULFATE ION | | Authors: | Kumari, P, Bhavesh, N.S. | | Deposit date: | 2019-11-23 | | Release date: | 2020-12-02 | | Last modified: | 2021-06-02 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Human DND1-RRM2 forms a non-canonical domain swapped dimer.
Protein Sci., 30, 2021
|
|
4IFQ
 
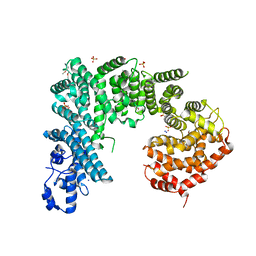 | | Crystal structure of Saccharomyces cerevisiae NUP192, residues 2 to 960 [ScNup192(2-960)] | | Descriptor: | IODIDE ION, Nucleoporin NUP192, SULFATE ION | | Authors: | Sampathkumar, P, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC), Nucleocytoplasmic Transport: a Target for Cellular Control (NPCXstals) | | Deposit date: | 2012-12-14 | | Release date: | 2013-02-20 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | Structure, dynamics, evolution, and function of a major scaffold component in the nuclear pore complex.
Structure, 21, 2013
|
|
3N7C
 
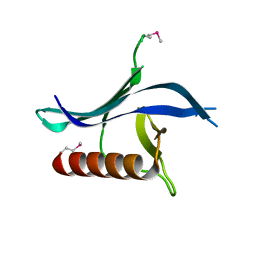 | | Crystal structure of the RAN binding domain from the nuclear pore complex component NUP2 from Ashbya gossypii | | Descriptor: | ABR034Wp | | Authors: | Sampathkumar, P, Manglicmot, D, Gilmore, J, Bain, K, Gheyi, T, Atwell, S, Thompson, D.A, Emtage, J.S, Wasserman, S, Sauder, J.M, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2010-05-26 | | Release date: | 2010-06-16 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Crystal structure of the RAN binding domain from the nuclear pore complex component NUP2 from Ashbya gossypii
To be Published
|
|
3NO8
 
 | | Crystal structure of the PHR domain from human BTBD2 Protein | | Descriptor: | BTB/POZ domain-containing protein 2, GLYCEROL, SULFATE ION | | Authors: | Sampathkumar, P, Miller, S, Rutter, M, Bain, K, Gheyi, T, Atwell, S, Thompson, D.A, Emtage, J.S, Wasserman, S, Sauder, J.M, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2010-06-24 | | Release date: | 2010-08-25 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of the PHR domain from human BTBD2 Protein
To be Published
|
|
3KES
 
 | | Crystal structure of the autoproteolytic domain from the nuclear pore complex component NUP145 from Saccharomyces cerevisiae in the Hexagonal, P61 space group | | Descriptor: | 1,2-ETHANEDIOL, Nucleoporin NUP145 | | Authors: | Sampathkumar, P, Ozyurt, S.A, Do, J, Bain, K, Dickey, M, Gheyi, T, Sali, A, Kim, S.J, Phillips, J, Pieper, U, Fernandez-Martinez, J, Franke, J.D, Atwell, S, Thompson, D.A, Emtage, J.S, Wasserman, S, Rout, M, Sauder, J.M, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-10-26 | | Release date: | 2009-12-22 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structures of the autoproteolytic domain from the Saccharomyces cerevisiae nuclear pore complex component, Nup145.
Proteins, 78, 2010
|
|
3JTY
 
 | | Crystal structure of a BenF-like porin from Pseudomonas fluorescens Pf-5 | | Descriptor: | BenF-like porin, LAURYL DIMETHYLAMINE-N-OXIDE | | Authors: | Sampathkumar, P, Lu, F, Zhao, X, Wasserman, S, Iuzuka, M, Bain, K, Rutter, M, Gheyi, T, Atwell, S, Luz, J, Gilmore, J, Sauder, J.M, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-09-14 | | Release date: | 2009-10-20 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.58 Å) | | Cite: | Structure of a putative BenF-like porin from Pseudomonas fluorescens Pf-5 at 2.6 A resolution.
Proteins, 78, 2010
|
|
3P3D
 
 | | Crystal structure of the Nup53 RRM domain from Pichia guilliermondii | | Descriptor: | Nucleoporin 53 | | Authors: | Sampathkumar, P, Shawn, C, Bain, K, Gilmore, J, Gheyi, T, Atwell, S, Thompson, D.A, Emtage, J.S, Wasserman, S, Sauder, J.M, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2010-10-04 | | Release date: | 2011-01-19 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Crystal structure of the Nup53 RRM domain from Pichia guilliermondii
To be Published
|
|
3KFO
 
 | | Crystal structure of the C-terminal domain from the nuclear pore complex component NUP133 from Saccharomyces cerevisiae | | Descriptor: | GLYCEROL, Nucleoporin NUP133 | | Authors: | Sampathkumar, P, Bonanno, J.B, Miller, S, Bain, K, Dickey, M, Gheyi, T, Almo, S.C, Rout, M, Sali, A, Phillips, J, Pieper, U, Fernandez-Martinez, J, Franke, J.D, Atwell, S, Thompson, D.A, Emtage, J.S, Wasserman, S, Sauder, J.M, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-10-27 | | Release date: | 2010-01-26 | | Last modified: | 2021-02-10 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of the C-terminal domain of Saccharomyces cerevisiae Nup133, a component of the nuclear pore complex.
Proteins, 79, 2011
|
|
3HWJ
 
 | | Crystal structure of the second PHR domain of Mouse Myc-binding protein 2 (MYCBP-2) | | Descriptor: | DIMETHYL SULFOXIDE, E3 ubiquitin-protein ligase MYCBP2 | | Authors: | Sampathkumar, P, Ozyurt, S.A, Wasserman, S.R, Miller, S.A, Bain, K.T, Rutter, M.E, Gheyi, T, Klemke, R.L, Atwell, S, Sauder, J.M, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-06-17 | | Release date: | 2009-07-21 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structures of PHR domains from Mus musculus Phr1 (Mycbp2) explain the loss-of-function mutation (Gly1092-->Glu) of the C. elegans ortholog RPM-1.
J.Mol.Biol., 397, 2010
|
|
3NF5
 
 | | Crystal structure of the C-terminal domain of nuclear pore complex component NUP116 from Candida glabrata | | Descriptor: | GLYCEROL, Nucleoporin NUP116 | | Authors: | Sampathkumar, P, Manglicmot, D, Bain, K, Gilmore, J, Gheyi, T, Rout, M, Sali, A, Atwell, S, Thompson, D.A, Emtage, J.S, Wasserman, S, Sauder, J.M, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2010-06-09 | | Release date: | 2010-08-04 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Atomic structure of the nuclear pore complex targeting domain of a Nup116 homologue from the yeast, Candida glabrata.
Proteins, 80, 2012
|
|
3KEP
 
 | | Crystal structure of the autoproteolytic domain from the nuclear pore complex component NUP145 from Saccharomyces cerevisiae | | Descriptor: | 1,2-ETHANEDIOL, Nucleoporin NUP145 | | Authors: | Sampathkumar, P, Ozyurt, S.A, Do, J, Bain, K, Dickey, M, Gheyi, T, Sali, A, Kim, S.J, Phillips, J, Pieper, U, Fernandez-Martinez, J, Franke, J.D, Atwell, S, Thompson, D.A, Emtage, J.S, Wasserman, S, Rout, M, Sauder, J.M, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-10-26 | | Release date: | 2009-12-22 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Structures of the autoproteolytic domain from the Saccharomyces cerevisiae nuclear pore complex component, Nup145.
Proteins, 78, 2010
|
|
3PVE
 
 | | Crystal structure of the G2 domain of Agrin from Mus Musculus | | Descriptor: | Agrin, Agrin protein | | Authors: | Sampathkumar, P, Do, J, Bain, K, Freeman, J, Gheyi, T, Atwell, S, Thompson, D.A, Emtage, J.S, Wasserman, S, Sauder, J.M, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2010-12-07 | | Release date: | 2011-01-19 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal structure of the G2 domain of Agrin from Mus Musculus
To be Published
|
|
4ESK
 
 | |
4ETY
 
 | |
4EJM
 
 | |
4EJ6
 
 | |
3CVL
 
 | | Structure of Peroxisomal Targeting Signal 1 (PTS1) binding domain of Trypanosoma brucei Peroxin 5 (TbPEX5)complexed to T. brucei Phosphofructokinase (PFK) PTS1 peptide | | Descriptor: | Peroxisome targeting signal 1 receptor PEX5, T. brucei PFK PTS1 peptide Ac-HEELAKL | | Authors: | Sampathkumar, P, Roach, C, Michels, P.A.M, Hol, W.G.J. | | Deposit date: | 2008-04-18 | | Release date: | 2008-06-24 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structural Insights into the recognition of peroxisomal targeting signal 1 by Trypanosoma brucei peroxin 5.
J.Mol.Biol., 381, 2008
|
|
3CVQ
 
 | | Structure of Peroxisomal Targeting Signal 1 (PTS1) binding domain of Trypanosoma brucei Peroxin 5 (TbPEX5)complexed to PTS1 peptide (7-SKL) | | Descriptor: | GLYCEROL, PTS1 peptide 7-SKL (Ac-SNRWSKL), Peroxisome targeting signal 1 receptor PEX5 | | Authors: | Sampathkumar, P, Roach, C, Michels, P.A.M, Hol, W.G.J. | | Deposit date: | 2008-04-18 | | Release date: | 2008-06-24 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3.01 Å) | | Cite: | Structural Insights into the recognition of peroxisomal targeting signal 1 by Trypanosoma brucei peroxin 5.
J.Mol.Biol., 381, 2008
|
|
3CVN
 
 | | Structure of Peroxisomal Targeting Signal 1 (PTS1) binding domain of Trypanosoma brucei Peroxin 5 (TbPEX5)complexed to T. brucei Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) PTS1 peptide | | Descriptor: | 1,2-ETHANEDIOL, Peroxisome targeting signal 1 receptor, T. brucei GAPDH PTS1 peptide Ac-DRDAAKL | | Authors: | Sampathkumar, P, Roach, C, Michels, P.A.M, Hol, W.G.J. | | Deposit date: | 2008-04-18 | | Release date: | 2008-06-24 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Insights into the recognition of peroxisomal targeting signal 1 by Trypanosoma brucei peroxin 5.
J.Mol.Biol., 381, 2008
|
|
3CVP
 
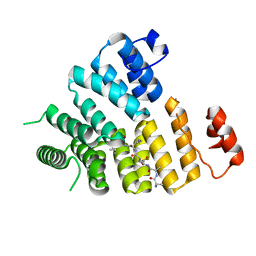 | | Structure of Peroxisomal Targeting Signal 1 (PTS1) binding domain of Trypanosoma brucei Peroxin 5 (TbPEX5)complexed to PTS1 peptide (10-SKL) | | Descriptor: | 10-SKL PTS1 peptide Ac-GTLSNRASKL, Peroxisome targeting signal 1 receptor PEX5 | | Authors: | Sampathkumar, P, Roach, C, Michels, P.A.M, Hol, W.G.J. | | Deposit date: | 2008-04-18 | | Release date: | 2008-06-24 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Insights into the recognition of peroxisomal targeting signal 1 by Trypanosoma brucei peroxin 5.
J.Mol.Biol., 381, 2008
|
|
