2PC8
 
 | | E292Q mutant of EXO-B-(1,3)-Glucanase from Candida Albicans in complex with two separately bound glucopyranoside units at 1.8 A | | Descriptor: | Hypothetical protein XOG1, beta-D-glucopyranose | | Authors: | Cutfield, S.M, Cutfield, J.F, Patrick, W.M. | | Deposit date: | 2007-03-29 | | Release date: | 2008-04-01 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Carbohydrate binding sites in Candida albicans exo-beta-1,3-glucanase and the role of the Phe-Phe 'clamp' at the active site entrance.
Febs J., 277, 2010
|
|
2PBO
 
 | |
4ALD
 
 | | HUMAN MUSCLE FRUCTOSE 1,6-BISPHOSPHATE ALDOLASE COMPLEXED WITH FRUCTOSE 1,6-BISPHOSPHATE | | Descriptor: | 1,6-FRUCTOSE DIPHOSPHATE (LINEAR FORM), FRUCTOSE-BISPHOSPHATE ALDOLASE | | Authors: | Dalby, A.R, Dauter, Z, Littlechild, J.A. | | Deposit date: | 1998-07-26 | | Release date: | 1999-03-02 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of human muscle aldolase complexed with fructose 1,6-bisphosphate: mechanistic implications.
Protein Sci., 8, 1999
|
|
1IGC
 
 | |
1O8P
 
 | | Unbound structure of CsCBM6-3 from Clostridium stercorarium | | Descriptor: | CALCIUM ION, PUTATUVE ENDO-XYLANASE | | Authors: | Boraston, A.B, Notenboom, V, Warren, R.A.J, Kilbrun, D.G, Rose, D.R, Davies, G.J. | | Deposit date: | 2002-11-28 | | Release date: | 2003-03-13 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure and ligand binding of carbohydrate-binding module CsCBM6-3 reveals similarities with fucose-specific lectins and "galactose-binding" domains.
J. Mol. Biol., 327, 2003
|
|
5NO7
 
 | | Crystal Structure of a Xylan-active Lytic Polysaccharide Monooxygenase from Pycnoporus coccineus. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Lytic polysaccharide monooxygenase, SULFATE ION, ... | | Authors: | Ladeveze, S, Couturier, M, Sulzenbacher, G, Berrin, J.-G. | | Deposit date: | 2017-04-11 | | Release date: | 2018-01-17 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (3.01 Å) | | Cite: | Lytic xylan oxidases from wood-decay fungi unlock biomass degradation.
Nat. Chem. Biol., 14, 2018
|
|
1QGQ
 
 | | UDP-MANGANESE COMPLEX OF SPSA FROM BACILLUS SUBTILIS | | Descriptor: | GLYCEROL, MAGNESIUM ION, MANGANESE (II) ION, ... | | Authors: | Charnock, S.J. | | Deposit date: | 1999-05-04 | | Release date: | 2000-05-04 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure of the nucleotide-diphospho-sugar transferase, SpsA from Bacillus subtilis, in native and nucleotide-complexed forms.
Biochemistry, 38, 1999
|
|
1QG8
 
 | | NATIVE (MAGNESIUM-CONTAINING) SPSA FROM BACILLUS SUBTILIS | | Descriptor: | GLYCEROL, MAGNESIUM ION, PROTEIN (SPORE COAT POLYSACCHARIDE BIOSYNTHESIS PROTEIN SPSA) | | Authors: | Charnock, S.J. | | Deposit date: | 1999-04-21 | | Release date: | 2000-04-21 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure of the nucleotide-diphospho-sugar transferase, SpsA from Bacillus subtilis, in native and nucleotide-complexed forms.
Biochemistry, 38, 1999
|
|
1QGS
 
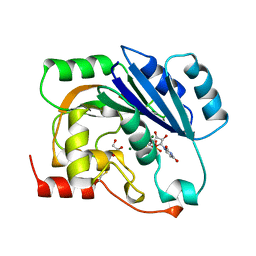 | | UDP-MAGNESIUM COMPLEX OF SPSA FROM BACILLUS SUBTILIS | | Descriptor: | GLYCEROL, MAGNESIUM ION, PROTEIN (SPORE COAT POLYSACCHARIDE BIOSYNTHESIS PROTEIN SPSA), ... | | Authors: | Charnock, S.J. | | Deposit date: | 1999-05-04 | | Release date: | 2000-05-04 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of the nucleotide-diphospho-sugar transferase, SpsA from Bacillus subtilis, in native and nucleotide-complexed forms.
Biochemistry, 38, 1999
|
|
258D
 
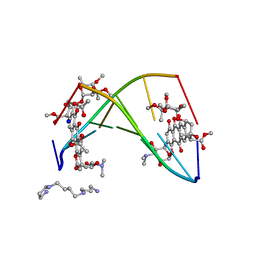 | | FACTORS AFFECTING SEQUENCE SELECTIVITY ON NOGALAMYCIN INTERCALATION: THE CRYSTAL STRUCTURE OF D(TGTACA)-NOGALAMYCIN | | Descriptor: | ACETATE ION, DNA (5'-D(*TP*GP*TP*AP*CP*A)-3'), NOGALAMYCIN, ... | | Authors: | Smith, C.K, Brannigan, J.A, Moore, M.H. | | Deposit date: | 1996-05-12 | | Release date: | 1996-06-20 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Factors affecting DNA sequence selectivity of nogalamycin intercalation: the crystal structure of d(TGTACA)2-nogalamycin2.
J.Mol.Biol., 263, 1996
|
|
1U8X
 
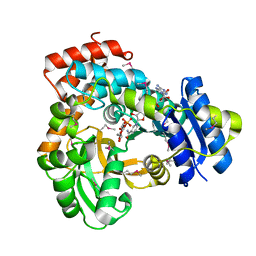 | | CRYSTAL STRUCTURE OF GLVA FROM BACILLUS SUBTILIS, A METAL-REQUIRING, NAD-DEPENDENT 6-PHOSPHO-ALPHA-GLUCOSIDASE | | Descriptor: | 6-O-phosphono-alpha-D-glucopyranose, MANGANESE (II) ION, Maltose-6'-phosphate glucosidase, ... | | Authors: | Rajan, S.S, Yang, X, Collart, F, Anderson, W.F, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2004-08-09 | | Release date: | 2004-08-24 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Novel Catalytic Mechanism of Glycoside Hydrolysis Based on the Structure of an NAD(+)/Mn(2+)-Dependent Phospho-alpha-Glucosidase from Bacillus subtilis.
STRUCTURE, 12, 2004
|
|
1HFU
 
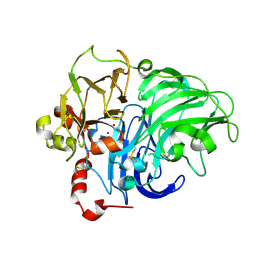 | | TYPE-2 CU-DEPLETED LACCASE FROM COPRINUS CINEREUS at 1.68 A resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, COPPER (II) ION, LACCASE 1, ... | | Authors: | Ducros, V, Brzozowski, A.M. | | Deposit date: | 2000-12-08 | | Release date: | 2001-12-06 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Structure of the Laccase from Coprinus Cinereus at 1.68A Resolution: Evidence for Different Type 2 Cu-Depleted Isoforms
Acta Crystallogr.,Sect.D, 57, 2001
|
|
5M77
 
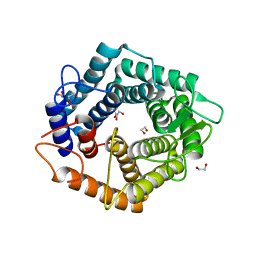 | | a GH76 family enzyme structure | | Descriptor: | 1,2-ETHANEDIOL, 1-thio-alpha-D-mannopyranose-(1-6)-[(3S,4R,5R)-4,5-dihydroxypiperidin-3-yl]methyl 1-thio-alpha-D-mannopyranoside, Alpha-1,6-mannanase | | Authors: | Jin, Y, Williams, S, Davies, G. | | Deposit date: | 2016-10-26 | | Release date: | 2017-08-09 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | An atypical interaction explains the high-affinity of a non-hydrolyzable S-linked 1,6-alpha-mannanase inhibitor.
Chem. Commun. (Camb.), 53, 2017
|
|
1EG1
 
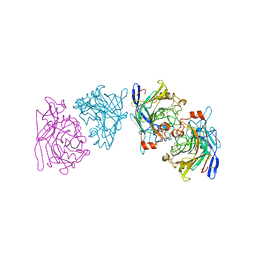 | | ENDOGLUCANASE I FROM TRICHODERMA REESEI | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ENDOGLUCANASE I | | Authors: | Kleywegt, G.J, Zou, J.-Y, Jones, T.A. | | Deposit date: | 1996-11-26 | | Release date: | 1997-08-20 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | The crystal structure of the catalytic core domain of endoglucanase I from Trichoderma reesei at 3.6 A resolution, and a comparison with related enzymes.
J.Mol.Biol., 272, 1997
|
|
5MQO
 
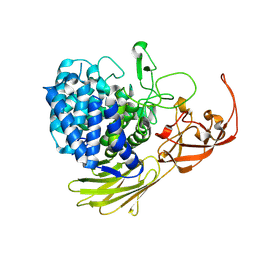 | | Glycoside hydrolase BT_1003 | | Descriptor: | Non-reducing end beta-L-arabinofuranosidase | | Authors: | Basle, A, Ndeh, D, Rogowski, A, Cartmell, A, Luis, A.S, Venditto, I, Labourel, A, Gilbert, H.J. | | Deposit date: | 2016-12-20 | | Release date: | 2017-03-22 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Complex pectin metabolism by gut bacteria reveals novel catalytic functions.
Nature, 544, 2017
|
|
5MQS
 
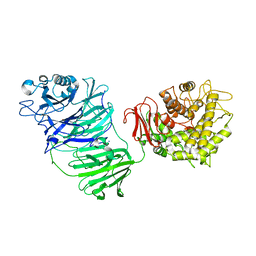 | | Sialidase BT_1020 | | Descriptor: | Beta-L-arabinobiosidase, CALCIUM ION, SODIUM ION, ... | | Authors: | Basle, A, Ndeh, D, Rogowski, A, Cartmell, A, Luis, A.S, Venditto, I, Labourel, A, Gilbert, H.J. | | Deposit date: | 2016-12-20 | | Release date: | 2017-03-22 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Complex pectin metabolism by gut bacteria reveals novel catalytic functions.
Nature, 544, 2017
|
|
5MQR
 
 | | Sialidase BT_1020 | | Descriptor: | 1,2-ETHANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Beta-L-arabinobiosidase, ... | | Authors: | Basle, A, Ndeh, D, Rogowski, A, Cartmell, A, Luis, A.S, Venditto, I, Labourel, A, Gilbert, H.J. | | Deposit date: | 2016-12-20 | | Release date: | 2017-03-22 | | Last modified: | 2017-08-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Complex pectin metabolism by gut bacteria reveals novel catalytic functions.
Nature, 544, 2017
|
|
5MQP
 
 | | Glycoside hydrolase BT_1002 | | Descriptor: | CALCIUM ION, Glycoside hydrolase BT_1002 | | Authors: | Basle, A, Ndeh, D, Rogowski, A, Cartmell, A, Luis, A.S, Venditto, I, Labourel, A, Gilbert, H.J. | | Deposit date: | 2016-12-20 | | Release date: | 2017-04-05 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The most complex carbohydrate known is degraded in the human gut by single organisms and not bacterial consortia
To Be Published
|
|
5MT2
 
 | | Glycoside hydrolase BT_0996 | | Descriptor: | Beta-galactosidase, GLYCEROL | | Authors: | Basle, A, Ndeh, D, Rogowski, A, Cartmell, A, Luis, A.S, Venditto, I, Labourel, A, Gilbert, H.J. | | Deposit date: | 2017-01-06 | | Release date: | 2017-03-22 | | Last modified: | 2017-08-30 | | Method: | X-RAY DIFFRACTION (1.41 Å) | | Cite: | Complex pectin metabolism by gut bacteria reveals novel catalytic functions.
Nature, 544, 2017
|
|
5MUJ
 
 | | BT0996 RGII Chain B Complex | | Descriptor: | ACETATE ION, Beta-galactosidase, alpha-L-rhamnopyranose-(1-2)-[alpha-L-rhamnopyranose-(1-3)]alpha-L-arabinopyranose-(1-4)-[4-O-[(1R)-1-hydroxyethyl]-2-O-methyl-alpha-L-fucopyranose-(1-2)]beta-D-galactopyranose-(1-2)-alpha-D-aceric acid-(1-4)-alpha-L-rhamnopyranose-(1-3)-3-C-(hydroxylmethyl)-alpha-D-erythrofuranose | | Authors: | Cartmell, A, Basle, A, Ndeh, D, Luis, A.S, Venditto, I, Labourel, A, Rogowski, A, Gilbert, H.J. | | Deposit date: | 2017-01-13 | | Release date: | 2017-04-05 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.37 Å) | | Cite: | Complex pectin metabolism by gut bacteria reveals novel catalytic functions.
Nature, 544, 2017
|
|
5MSY
 
 | | Glycoside hydrolase BT_1012 | | Descriptor: | AMMONIA, Glycoside hydrolase, PHOSPHATE ION | | Authors: | Basle, A, Ndeh, D, Rogowski, A, Cartmell, A, Luis, A.S, Venditto, I, Labourel, A, Gilbert, H.J. | | Deposit date: | 2017-01-06 | | Release date: | 2017-03-22 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Complex pectin metabolism by gut bacteria reveals novel catalytic functions.
Nature, 544, 2017
|
|
5LW4
 
 | |
5MQM
 
 | | Glycoside hydrolase BT_0986 | | Descriptor: | CALCIUM ION, D-rhamnopyranose tetrazole, Glycosyl hydrolases family 2, ... | | Authors: | Basle, A, Ndeh, D, Rogowski, A, Cartmell, A, Luis, A.S, Venditto, I, Labourel, A, Gilbert, H.J. | | Deposit date: | 2016-12-20 | | Release date: | 2017-03-22 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Complex pectin metabolism by gut bacteria reveals novel catalytic functions.
Nature, 544, 2017
|
|
5MQN
 
 | | Glycoside hydrolase BT_0986 | | Descriptor: | CALCIUM ION, Glycosyl hydrolases family 2, sugar binding domain | | Authors: | Basle, A, Ndeh, D, Rogowski, A, Cartmell, A, Luis, A.S, Venditto, I, Labourel, A, Gilbert, H.J. | | Deposit date: | 2016-12-20 | | Release date: | 2017-03-22 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Complex pectin metabolism by gut bacteria reveals novel catalytic functions.
Nature, 544, 2017
|
|
5MSX
 
 | | Glycoside hydrolase BT_3662 | | Descriptor: | 1,2-ETHANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CALCIUM ION, ... | | Authors: | Basle, A, Ndeh, D, Rogowski, A, Cartmell, A, Luis, A.S, Venditto, I, Labourel, A, Gilbert, H.J. | | Deposit date: | 2017-01-06 | | Release date: | 2017-03-22 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Complex pectin metabolism by gut bacteria reveals novel catalytic functions.
Nature, 544, 2017
|
|
