7DWC
 
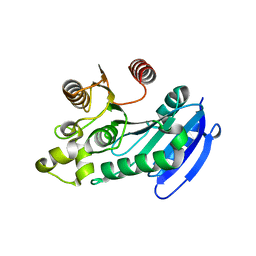 | | Bacteroides thetaiotaomicron VPI5482 BTAxe1 | | Descriptor: | Xylanase | | Authors: | Wang, L.Y, Wang, Y.L, Xin, F.J, Sun, L.C. | | Deposit date: | 2021-01-17 | | Release date: | 2022-01-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.804 Å) | | Cite: | Rational Design for Broadened Substrate Specificity and Enhanced Activity of a Novel Acetyl Xylan Esterase from Bacteroides thetaiotaomicron.
J.Agric.Food Chem., 69, 2021
|
|
3WVO
 
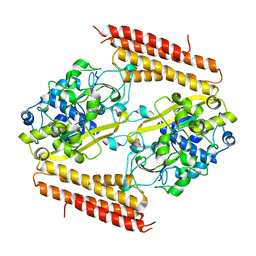 | | Crystal structure of Thermobifida fusca Cse1 | | Descriptor: | CRISPR-associated protein, Cse1 family | | Authors: | Yuan, Y.A, Tay, M. | | Deposit date: | 2014-06-02 | | Release date: | 2015-01-14 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.31 Å) | | Cite: | Crystal structure of Thermobifida fusca Cse1 reveals target DNA binding site.
Protein Sci., 24, 2015
|
|
7VY5
 
 | | Coxsackievirus B3 (VP3-234Q) incubation with CD55 at pH7.4 | | Descriptor: | Capsid protein VP1, Capsid protein VP2, Capsid protein VP3, ... | | Authors: | Wang, Q.L, Liu, C.C. | | Deposit date: | 2021-11-13 | | Release date: | 2022-01-19 | | Last modified: | 2022-08-10 | | Method: | ELECTRON MICROSCOPY (3.15 Å) | | Cite: | Molecular basis of differential receptor usage for naturally occurring CD55-binding and -nonbinding coxsackievirus B3 strains.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7VXZ
 
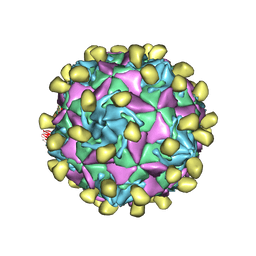 | |
7VYL
 
 | |
7VYK
 
 | |
7VXH
 
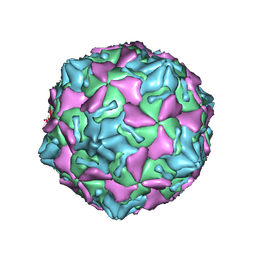 | | Coxsackievirus B3 full particle at pH7.4 (VP3-234Q) | | Descriptor: | Capsid protein VP1, Capsid protein VP2, Capsid protein VP3, ... | | Authors: | Wang, Q.L, Liu, C.C. | | Deposit date: | 2021-11-12 | | Release date: | 2022-01-19 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (2.95 Å) | | Cite: | Molecular basis of differential receptor usage for naturally occurring CD55-binding and -nonbinding coxsackievirus B3 strains.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7VY0
 
 | | Coxsackievirus B3 full particle at pH7.4 (VP3-234N) | | Descriptor: | Capsid protein VP1, Capsid protein VP2, Capsid protein VP3, ... | | Authors: | Wang, Q.L, Liu, C.C. | | Deposit date: | 2021-11-13 | | Release date: | 2022-01-19 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Molecular basis of differential receptor usage for naturally occurring CD55-binding and -nonbinding coxsackievirus B3 strains.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7VYM
 
 | |
7VY6
 
 | | Coxsackievirus B3(VP3-234N) incubate with CD55 at pH7.4 | | Descriptor: | Capsid protein VP1, Capsid protein VP2, Capsid protein VP3, ... | | Authors: | Wang, Q.L, Liu, C.C. | | Deposit date: | 2021-11-13 | | Release date: | 2022-01-19 | | Last modified: | 2022-08-10 | | Method: | ELECTRON MICROSCOPY (3.02 Å) | | Cite: | Molecular basis of differential receptor usage for naturally occurring CD55-binding and -nonbinding coxsackievirus B3 strains.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7W17
 
 | | Coxsackievirus B3 full particle at pH7.4 (VP3-234E) | | Descriptor: | PALMITIC ACID, VP1, VP2, ... | | Authors: | Wang, Q.L, Liu, C.C. | | Deposit date: | 2021-11-19 | | Release date: | 2022-01-19 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Molecular basis of differential receptor usage for naturally occurring CD55-binding and -nonbinding coxsackievirus B3 strains.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7W14
 
 | |
4GST
 
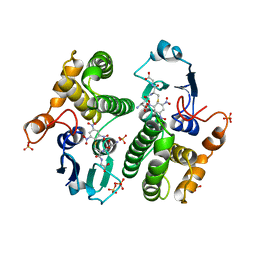 | | REACTION COORDINATE MOTION IN AN SNAR REACTION CATALYZED BY GLUTATHIONE TRANSFERASE | | Descriptor: | 1-(S-GLUTATHIONYL)-2,4,6-TRINITROCYCLOHEXA-2,5-DIENE, GLUTATHIONE S-TRANSFERASE, SULFATE ION | | Authors: | Ji, X, Armstrong, R.N, Gilliland, G.L. | | Deposit date: | 1993-07-20 | | Release date: | 1993-10-31 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Snapshots along the reaction coordinate of an SNAr reaction catalyzed by glutathione transferase.
Biochemistry, 32, 1993
|
|
2GST
 
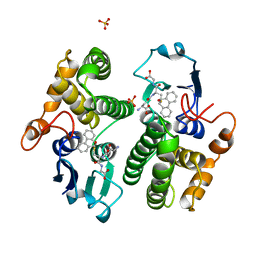 | | STRUCTURE OF THE XENOBIOTIC SUBSTRATE BINDING SITE OF A GLUTATHIONE S-TRANSFERASE AS REVEALED BY X-RAY CRYSTALLOGRAPHIC ANALYSIS OF PRODUCT COMPLEXES WITH THE DIASTEREOMERS OF 9-(S-GLUTATHIONYL)-10-HYDROXY-9, 10-DIHYDROPHENANTHRENE | | Descriptor: | GLUTATHIONE S-TRANSFERASE, L-gamma-glutamyl-S-[(9S,10S)-10-hydroxy-9,10-dihydrophenanthren-9-yl]-L-cysteinylglycine, SULFATE ION | | Authors: | Ji, X, Armstrong, R.N, Gilliland, G.L. | | Deposit date: | 1993-06-07 | | Release date: | 1993-10-31 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure and function of the xenobiotic substrate binding site of a glutathione S-transferase as revealed by X-ray crystallographic analysis of product complexes with the diastereomers of 9-(S-glutathionyl)-10-hydroxy-9,10-dihydrophenanthrene.
Biochemistry, 33, 1994
|
|
7E8C
 
 | | SARS-CoV-2 S-6P in complex with 9 Fabs | | Descriptor: | 368-2 H, 368-2 L, 604 H, ... | | Authors: | Du, S, Xiao, J, Zhang, Z. | | Deposit date: | 2021-03-01 | | Release date: | 2021-06-09 | | Last modified: | 2021-07-14 | | Method: | ELECTRON MICROSCOPY (3.16 Å) | | Cite: | Humoral immune response to circulating SARS-CoV-2 variants elicited by inactivated and RBD-subunit vaccines.
Cell Res., 31, 2021
|
|
7E8F
 
 | | SARS-CoV-2 NTD in complex with N9 Fab | | Descriptor: | 368-2 H, 368-2 L, 604 H, ... | | Authors: | Du, S, Xiao, J, Zhang, Z. | | Deposit date: | 2021-03-01 | | Release date: | 2021-06-09 | | Last modified: | 2021-07-14 | | Method: | ELECTRON MICROSCOPY (3.18 Å) | | Cite: | Humoral immune response to circulating SARS-CoV-2 variants elicited by inactivated and RBD-subunit vaccines.
Cell Res., 31, 2021
|
|
4TQ4
 
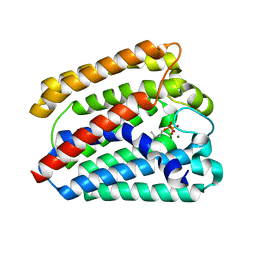 | | Structure of a UbiA homolog from Archaeoglobus fulgidus bound to DMAPP and Mg2+ | | Descriptor: | DIMETHYLALLYL DIPHOSPHATE, MAGNESIUM ION, prenyltransferase | | Authors: | Huang, H, Levin, E.J, Bai, Y, Zhou, M, New York Consortium on Membrane Protein Structure (NYCOMPS) | | Deposit date: | 2014-06-10 | | Release date: | 2014-07-16 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.5025 Å) | | Cite: | Structure of a Membrane-Embedded Prenyltransferase Homologous to UBIAD1.
Plos Biol., 12, 2014
|
|
4TQ6
 
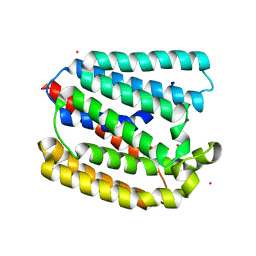 | | Structure of a UbiA homolog from Archaeoglobus fulgidus bound to Cd2+ | | Descriptor: | CADMIUM ION, prenyltransferase | | Authors: | Huang, H, Levin, E.J, Bai, Y, Zhou, M, New York Consortium on Membrane Protein Structure (NYCOMPS) | | Deposit date: | 2014-06-10 | | Release date: | 2014-07-16 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.0678 Å) | | Cite: | Structure of a Membrane-Embedded Prenyltransferase Homologous to UBIAD1.
Plos Biol., 12, 2014
|
|
4TQ5
 
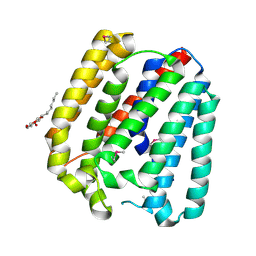 | | Structure of a UbiA homolog from Archaeoglobus fulgidus | | Descriptor: | octyl beta-D-glucopyranoside, prenyltransferase | | Authors: | Huang, H, Levin, E.J, Bai, Y, Zhou, M, New York Consortium on Membrane Protein Structure (NYCOMPS) | | Deposit date: | 2014-06-10 | | Release date: | 2014-07-16 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3.2023 Å) | | Cite: | Structure of a Membrane-Embedded Prenyltransferase Homologous to UBIAD1.
Plos Biol., 12, 2014
|
|
4TQ3
 
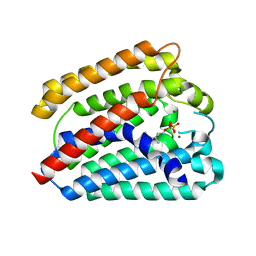 | | Structure of a UbiA homolog from Archaeoglobus fulgidus bound to GPP and Mg2+ | | Descriptor: | GERANYL DIPHOSPHATE, MAGNESIUM ION, Prenyltransferase | | Authors: | Huang, H, Levin, E.J, Bai, Y, Zhou, M, New York Consortium on Membrane Protein Structure (NYCOMPS) | | Deposit date: | 2014-06-10 | | Release date: | 2014-07-16 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.4076 Å) | | Cite: | Structure of a Membrane-Embedded Prenyltransferase Homologous to UBIAD1.
Plos Biol., 12, 2014
|
|
1ABN
 
 | |
6KUU
 
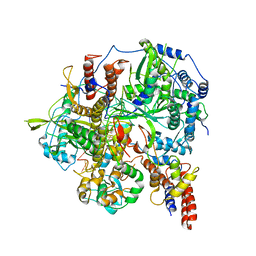 | | Structure of influenza D virus polymerase bound to vRNA promoter in Mode B conformation (Class B3) | | Descriptor: | 3'-vRNA, 5'-vRNA, Polymerase 3, ... | | Authors: | Peng, Q, Peng, R, Qi, J, Gao, G.F, Shi, Y. | | Deposit date: | 2019-09-02 | | Release date: | 2019-12-11 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structure of influenza D virus polymerase bound to vRNA promoter in Mode B conformation (Class B3)
To Be Published
|
|
7EW8
 
 | | Legionella pneumophila effector AnkD | | Descriptor: | ANK_REP_REGION domain-containing protein | | Authors: | Chen, T.T, Lin, Y.L. | | Deposit date: | 2021-05-24 | | Release date: | 2022-06-01 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.594 Å) | | Cite: | Atypical Legionella GTPase effector hijacks host vesicular transport factor p115 to regulate host lipid droplet.
Sci Adv, 8, 2022
|
|
7E7Y
 
 | |
7E7X
 
 | |
