4MQ6
 
 | |
4MUE
 
 | |
4MUF
 
 | |
5TJ3
 
 | | Crystal structure of wild type alkaline phosphatase PafA to 1.7A resolution | | Descriptor: | Alkaline phosphatase PafA, ZINC ION | | Authors: | Lyubimov, A.Y, Sunden, F, Ressl, S, Herschlag, D. | | Deposit date: | 2016-10-03 | | Release date: | 2016-11-16 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Mechanistic and Evolutionary Insights from Comparative Enzymology of Phosphomonoesterases and Phosphodiesterases across the Alkaline Phosphatase Superfamily.
J.Am.Chem.Soc., 138, 2016
|
|
4HB0
 
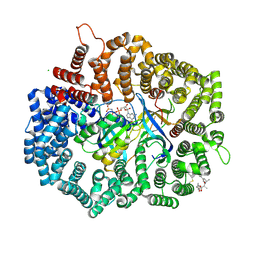 | | Crystal structure of CRM1 inhibitor Leptomycin B in complex with CRM1(K541Q,K542Q,R543S,K545Q,K548Q,K579Q)-Ran-RanBP1 | | Descriptor: | CHLORIDE ION, Exportin-1, GTP-binding nuclear protein Ran, ... | | Authors: | Sun, Q, Chook, Y.M. | | Deposit date: | 2012-09-27 | | Release date: | 2013-01-09 | | Last modified: | 2017-07-26 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Nuclear export inhibition through covalent conjugation and hydrolysis of Leptomycin B by CRM1.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4HAV
 
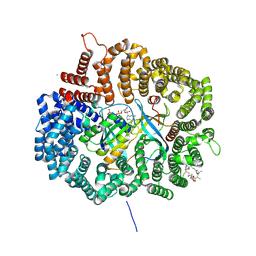 | |
4HAX
 
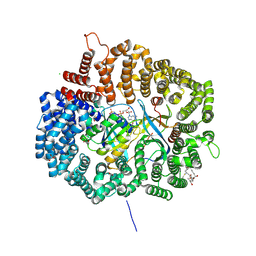 | |
4HB2
 
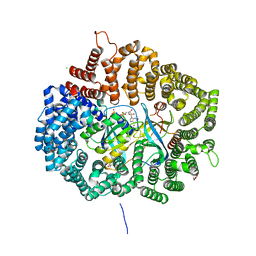 | | Crystal structure of CRM1-Ran-RanBP1 | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Exportin-1, ... | | Authors: | Sun, Q, Chook, Y.M. | | Deposit date: | 2012-09-27 | | Release date: | 2013-01-09 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Nuclear export inhibition through covalent conjugation and hydrolysis of Leptomycin B by CRM1.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4HAU
 
 | |
1M8Z
 
 | |
4PRU
 
 | | Crystal structure of dimethyllysine hen egg-white lysozyme in complex with sclx4 at 2.2 A resolution | | Descriptor: | 25,26,27,28-tetrahydroxypentacyclo[19.3.1.1~3,7~.1~9,13~.1~15,19~]octacosa-1(25),3(28),4,6,9(27),10,12,15(26),16,18,21,23-dodecaene-5,11,17,23-tetrasulfonic acid, GLYCEROL, Lysozyme C | | Authors: | McGovern, R.E, Lyons, J.A, Crowley, P.B. | | Deposit date: | 2014-03-06 | | Release date: | 2014-11-12 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural study of a small molecule receptor bound to dimethyllysine in lysozyme.
Chem Sci, 6, 2015
|
|
4HAT
 
 | |
4DF6
 
 | |
4I1R
 
 | | Human MALT1 (caspase-IG3) in complex with thioridazine | | Descriptor: | 10-{2-[(2S)-1-methylpiperidin-2-yl]ethyl}-2-(methylsulfanyl)-10H-phenothiazine, Mucosa-associated lymphoid tissue lymphoma translocation protein 1 | | Authors: | Schlauderer, F, Lammens, K, Hopfner, K.P. | | Deposit date: | 2012-11-21 | | Release date: | 2013-08-28 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural Analysis of Phenothiazine Derivatives as Allosteric Inhibitors of the MALT1 Paracaspase.
Angew.Chem.Int.Ed.Engl., 52, 2013
|
|
1AOD
 
 | | PHOSPHATIDYLINOSITOL-SPECIFIC PHOSPHOLIPASE C FROM LISTERIA MONOCYTOGENES | | Descriptor: | 1,2,3,4,5,6-HEXAHYDROXY-CYCLOHEXANE, PHOSPHATIDYLINOSITOL-SPECIFIC PHOSPHOLIPASE C | | Authors: | Heinz, D.W, Moser, J. | | Deposit date: | 1997-07-02 | | Release date: | 1998-01-07 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of the phosphatidylinositol-specific phospholipase C from the human pathogen Listeria monocytogenes.
J.Mol.Biol., 273, 1997
|
|
1DSP
 
 | | CYTOCHROME C PEROXIDASE H175G MUTANT, IMIDAZOLE COMPLEX AT PH 7, ROOM TEMPERATURE. | | Descriptor: | CYTOCHROME C PEROXIDASE, IMIDAZOLE, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Hirst, J, Wilcox, S.K, Williams, P.A, McRee, D.E, Goodin, D.B. | | Deposit date: | 2000-01-07 | | Release date: | 2001-03-07 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Replacement of the axial histidine ligand with imidazole in cytochrome c peroxidase. 1. Effects on structure.
Biochemistry, 40, 2001
|
|
1DSG
 
 | | CYTOCHROME C PEROXIDASE H175G MUTANT, IMIDAZOLE COMPLEX AT PH 5, ROOM TEMPERATURE. | | Descriptor: | CYTOCHROME C PEROXIDASE, IMIDAZOLE, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Hirst, J, Wilcox, S.K, Williams, P.A, McRee, D.E, Goodin, D.B. | | Deposit date: | 2000-01-07 | | Release date: | 2001-03-07 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.56 Å) | | Cite: | Replacement of the axial histidine ligand with imidazole in cytochrome c peroxidase. 1. Effects on structure.
Biochemistry, 40, 2001
|
|
1DS4
 
 | | CYTOCHROME C PEROXIDASE H175G MUTANT, IMIDAZOLE COMPLEX, PH 6, 100K | | Descriptor: | CYTOCHROME C PEROXIDASE, IMIDAZOLE, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Hirst, J, Wilcox, S.K, Williams, P.A, McRee, D.E, Goodin, D.B. | | Deposit date: | 2000-01-07 | | Release date: | 2001-03-07 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Replacement of the axial histidine ligand with imidazole in cytochrome c peroxidase. 1. Effects on structure.
Biochemistry, 40, 2001
|
|
1DSE
 
 | | CYTOCHROME C PEROXIDASE H175G MUTANT, IMIDAZOLE COMPLEX, WITH PHOSPHATE BOUND, PH 6, 100K | | Descriptor: | CYTOCHROME C PEROXIDASE, IMIDAZOLE, PHOSPHATE ION, ... | | Authors: | Hirst, J, Wilcox, S.K, Williams, P.A, McRee, D.E, Goodin, D.B. | | Deposit date: | 2000-01-07 | | Release date: | 2001-03-07 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Replacement of the axial histidine ligand with imidazole in cytochrome c peroxidase. 1. Effects on structure.
Biochemistry, 40, 2001
|
|
1DSO
 
 | | CYTOCHROME C PEROXIDASE H175G MUTANT, IMIDAZOLE COMPLEX AT PH 6, ROOM TEMPERATURE. | | Descriptor: | CYTOCHROME C PEROXIDASE, IMIDAZOLE, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Hirst, J, Wilcox, S.K, Williams, P.A, McRee, D.E, Goodin, D.B. | | Deposit date: | 2000-01-07 | | Release date: | 2001-03-07 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Replacement of the axial histidine ligand with imidazole in cytochrome c peroxidase. 1. Effects on structure.
Biochemistry, 40, 2001
|
|
5OT9
 
 | |
5F0V
 
 | | X-ray crystal structure of a thiolase from Escherichia coli at 1.8 A resolution | | Descriptor: | 1,2-ETHANEDIOL, Acetyl-CoA acetyltransferase | | Authors: | Ithayaraja, M, Neelanjana, J, Wierenga, R, Savithri, H.S, Murthy, M.R.N. | | Deposit date: | 2015-11-28 | | Release date: | 2016-07-13 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of a thiolase from Escherichia coli at 1.8 angstrom resolution.
Acta Crystallogr.,Sect.F, 72, 2016
|
|
5F38
 
 | | X-ray crystal structure of a thiolase from Escherichia coli at 1.8 A resolution | | Descriptor: | 1,2-ETHANEDIOL, Acetyl-CoA acetyltransferase, COENZYME A, ... | | Authors: | Ithayaraja, M, Neelanjana, J, Wierenga, R, Savithri, H.S, Murthy, M.R.N. | | Deposit date: | 2015-12-02 | | Release date: | 2016-07-13 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of a thiolase from Escherichia coli at 1.8 angstrom resolution.
Acta Crystallogr.,Sect.F, 72, 2016
|
|
1RSG
 
 | |
1TB4
 
 | |
