4LED
 
 | | The Crystal Structure of Pyocin L1 bound to D-rhamnose at 2.37 Angstroms | | Descriptor: | Pyocin L1, alpha-D-rhamnopyranose | | Authors: | Grinter, R, Roszak, A.W, Mccaughey, L, Cogdell, C.J, Walker, D. | | Deposit date: | 2013-06-25 | | Release date: | 2014-02-19 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | Lectin-Like Bacteriocins from Pseudomonas spp. Utilise D-Rhamnose Containing Lipopolysaccharide as a Cellular Receptor.
Plos Pathog., 10, 2014
|
|
4LE7
 
 | | The Crystal Structure of Pyocin L1 at 2.09 Angstroms | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Pyocin L1 | | Authors: | Grinter, R, Roszak, A.W, Mccaughey, L, Cogdell, R.J, Walker, D. | | Deposit date: | 2013-06-25 | | Release date: | 2014-02-19 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Lectin-Like Bacteriocins from Pseudomonas spp. Utilise D-Rhamnose Containing Lipopolysaccharide as a Cellular Receptor.
Plos Pathog., 10, 2014
|
|
4LEA
 
 | | The Crystal Structure of Pyocin L1 bound to D-mannose at 2.55 Angstroms | | Descriptor: | Pyocin L1, beta-D-mannopyranose | | Authors: | Grinter, R, Roszak, A.W, Mccaughey, L, Cogdell, C.J, Walker, D. | | Deposit date: | 2013-06-25 | | Release date: | 2014-02-19 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Lectin-Like Bacteriocins from Pseudomonas spp. Utilise D-Rhamnose Containing Lipopolysaccharide as a Cellular Receptor.
Plos Pathog., 10, 2014
|
|
4N58
 
 | | Crystal Structure of Pectocin M2 at 1.86 Angstroms | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CHLORIDE ION, FE2/S2 (INORGANIC) CLUSTER, ... | | Authors: | Grinter, R, Roszak, A.W, Zeth, K, Cogdell, C.J, Walker, D. | | Deposit date: | 2013-10-09 | | Release date: | 2014-06-04 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Structure of the atypical bacteriocin pectocin M2 implies a novel mechanism of protein uptake.
Mol.Microbiol., 93, 2014
|
|
4N59
 
 | | The Crystal Structure of Pectocin M2 at 2.3 Angstroms | | Descriptor: | CHLORIDE ION, FE2/S2 (INORGANIC) CLUSTER, Pectocin M2, ... | | Authors: | Zeth, K, Grinter, R, Roszak, A.W, Cogdell, R.J, Walker, D. | | Deposit date: | 2013-10-09 | | Release date: | 2014-06-04 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of the atypical bacteriocin pectocin M2 implies a novel mechanism of protein uptake.
Mol.Microbiol., 93, 2014
|
|
1GQO
 
 | |
6HN7
 
 | | Hijacking the Hijackers: Escherichia coli Pathogenicity Islands Redirect Helper Phage Packaging for Their Own Benefit. | | Descriptor: | Redirecting phage packaging protein C (RppC), Terminase small subunit | | Authors: | Penades, J.R, Bacarizo, J, Marina, A, Alqasmi, M, Fillol-Salom, A, Roszak, A.W, Ciges-Tomas, J.R. | | Deposit date: | 2018-09-14 | | Release date: | 2019-07-31 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Hijacking the Hijackers: Escherichia coli Pathogenicity Islands Redirect Helper Phage Packaging for Their Own Benefit.
Mol.Cell, 75, 2019
|
|
6HRN
 
 | | C-Phycocyanin from heterocyst forming filamentous cyanobacterium Nostoc sp. WR13 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Alpha Subunit of Cyanobacterial Phycocyanin protein, Beta Subunit of Cyanobacterial Phycocyanin protein, ... | | Authors: | Patel, H.M, Roszak, A.W, Madamwar, D, Cogdell, R.J. | | Deposit date: | 2018-09-27 | | Release date: | 2019-06-05 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.513 Å) | | Cite: | Crystal structure of phycocyanin from heterocyst-forming filamentous cyanobacterium Nostoc sp. WR13.
Int.J.Biol.Macromol., 135, 2019
|
|
6HLK
 
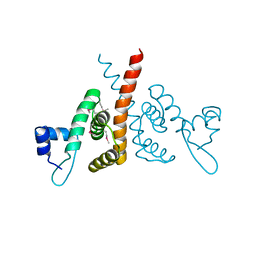 | | Hijacking the Hijackers: Escherichia coli Pathogenicity Islands Redirect Helper Phage Packaging for Their Own Benefit. | | Descriptor: | Redirecting phage packaging protein C (RppC) | | Authors: | Penades, J.R, Bacarizo, J, Marina, A, Alqasmi, M, Fillol-Salom, A, Roszak, A.W, Ciges-Tomas, J.R. | | Deposit date: | 2018-09-11 | | Release date: | 2019-07-31 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | Hijacking the Hijackers: Escherichia coli Pathogenicity Islands Redirect Helper Phage Packaging for Their Own Benefit.
Mol.Cell, 75, 2019
|
|
3TG7
 
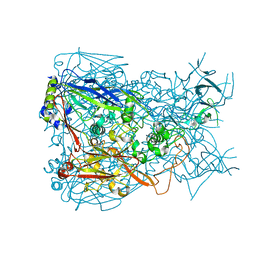 | | Crystal structure of Adenovirus serotype 5 hexon at 1.6A resolution | | Descriptor: | Hexon protein | | Authors: | Zhu, Y, Roszak, A.W, Isaacs, N.W, McVey, J.H, Nicklin, S.A, Baker, A.H. | | Deposit date: | 2011-08-17 | | Release date: | 2011-11-02 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | crystal structure of Adenovirus serotype 5 hexon at 1.6A resolution
To be Published
|
|
3TJB
 
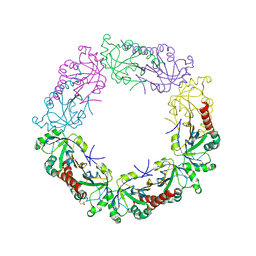 | | Crystal structure of wild-type human peroxiredoxin IV | | Descriptor: | Peroxiredoxin-4 | | Authors: | Cao, Z, Tavender, T.J, Roszak, A.W, Cogdell, R.J, Bulleid, N.J. | | Deposit date: | 2011-08-24 | | Release date: | 2011-10-12 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Crystal Structure of Reduced and of Oxidized Peroxiredoxin IV Enzyme Reveals a Stable Oxidized Decamer and a Non-disulfide-bonded Intermediate in the Catalytic Cycle.
J.Biol.Chem., 286, 2011
|
|
3TJJ
 
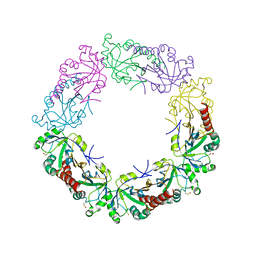 | | Crystal structure of human peroxiredoxin IV C245A mutant in sulfenylated form | | Descriptor: | Peroxiredoxin-4 | | Authors: | Cao, Z, Tavender, T.J, Roszak, A.W, Cogdell, R.J, Bulleid, N.J. | | Deposit date: | 2011-08-24 | | Release date: | 2011-10-12 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Crystal Structure of Reduced and of Oxidized Peroxiredoxin IV Enzyme Reveals a Stable Oxidized Decamer and a Non-disulfide-bonded Intermediate in the Catalytic Cycle.
J.Biol.Chem., 286, 2011
|
|
3TJF
 
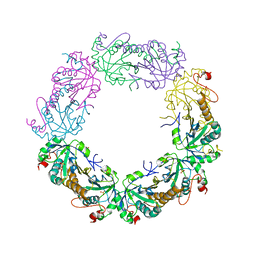 | | Crystal Structure of human peroxiredoxin IV C51A mutant in reduced form | | Descriptor: | Peroxiredoxin-4, SULFATE ION | | Authors: | Cao, Z, Tavender, T.J, Roszak, A.W, Cogdell, R.J, Bulleid, N.J. | | Deposit date: | 2011-08-24 | | Release date: | 2011-10-12 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Crystal Structure of Reduced and of Oxidized Peroxiredoxin IV Enzyme Reveals a Stable Oxidized Decamer and a Non-disulfide-bonded Intermediate in the Catalytic Cycle.
J.Biol.Chem., 286, 2011
|
|
3TJK
 
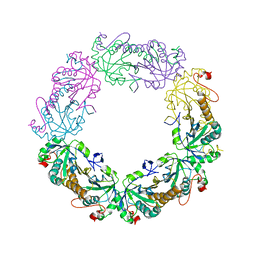 | | Crystal Structure of human peroxiredoxin IV C245A mutant in reduced form | | Descriptor: | Peroxiredoxin-4 | | Authors: | Cao, Z, Tavender, T.J, Roszak, A.W, Cogdell, R.J, Bulleid, N.J. | | Deposit date: | 2011-08-24 | | Release date: | 2011-10-12 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Crystal Structure of Reduced and of Oxidized Peroxiredoxin IV Enzyme Reveals a Stable Oxidized Decamer and a Non-disulfide-bonded Intermediate in the Catalytic Cycle.
J.Biol.Chem., 286, 2011
|
|
3TJG
 
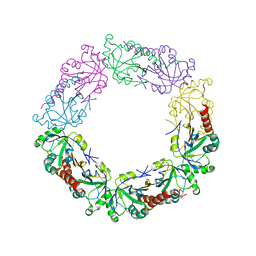 | | Crystal Structure of human peroxiredoxin IV C51A mutant in oxidized form | | Descriptor: | Peroxiredoxin-4 | | Authors: | Cao, Z, Tavender, T.J, Roszak, A.W, Cogdell, R.J, Bulleid, N.J. | | Deposit date: | 2011-08-24 | | Release date: | 2011-10-12 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Crystal Structure of Reduced and of Oxidized Peroxiredoxin IV Enzyme Reveals a Stable Oxidized Decamer and a Non-disulfide-bonded Intermediate in the Catalytic Cycle.
J.Biol.Chem., 286, 2011
|
|
1YIV
 
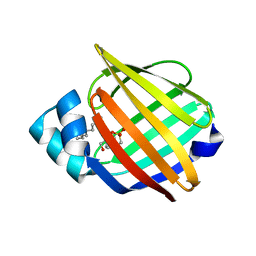 | | Structure of myelin P2 protein from Equine spinal cord | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, LAURYL DIMETHYLAMINE-N-OXIDE, Myelin P2 protein | | Authors: | Hunter, D.J.B, MacMaster, R, Rozak, A.W, Riboldi-Tunnicliffe, A, Grifiths, I.R, Freer, A.A. | | Deposit date: | 2005-01-13 | | Release date: | 2005-07-26 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of myelin P2 protein from equine spinal cord.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
4UHP
 
 | | Crystal structure of the pyocin AP41 DNase-Immunity complex | | Descriptor: | BACTERIOCIN IMMUNITY PROTEIN, LARGE COMPONENT OF PYOCIN AP41 | | Authors: | Joshi, A, Chen, S, Wojdyla, J.A, Kaminska, R, Kleanthous, C. | | Deposit date: | 2015-03-25 | | Release date: | 2015-08-05 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structures of the Ultra-High Affinity Protein-Protein Complexes of Pyocins S2 and Ap41 and Their Cognate Immunity Proteins from Pseudomonas Aeruginosa
J.Mol.Biol., 427, 2015
|
|
4UHQ
 
 | | Crystal structure of the pyocin AP41 DNase | | Descriptor: | CITRIC ACID, LARGE COMPONENT OF PYOCIN AP41, NICKEL (II) ION | | Authors: | Joshi, A, Chen, S, Wojdyla, J.A, Kaminska, R, Kleanthous, C. | | Deposit date: | 2015-03-25 | | Release date: | 2015-08-05 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structures of the Ultra-High Affinity Protein-Protein Complexes of Pyocins S2 and Ap41 and Their Cognate Immunity Proteins from Pseudomonas Aeruginosa
J.Mol.Biol., 427, 2015
|
|
2BT4
 
 | | Type II Dehydroquinase inhibitor complex | | Descriptor: | (1S,3R,4R,5S)-1,3,4-TRIHYDROXY-5-(3-PHENOXYPROPYL)CYCLOHEXANECARBOXYLIC ACID, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 3-DEHYDROQUINATE DEHYDRATASE, ... | | Authors: | Toscano, M.D, Stewart, K.A, Coggins, J.R, Lapthorn, A.J, Abell, C. | | Deposit date: | 2005-05-26 | | Release date: | 2006-02-13 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Rational Design of New Bifunctional Inhibitors of Type II Dehydroquinase.
Org.Biomol.Chem., 3, 2005
|
|
7ZDI
 
 | | PucB-LH2 complex from Rps. palustris | | Descriptor: | 1,2-Dihydro-psi,psi-caroten-1-ol, BACTERIOCHLOROPHYLL A, Light-harvesting protein, ... | | Authors: | Qian, P, Cogdell, R.J, Nguyen-Phan, T.C. | | Deposit date: | 2022-03-29 | | Release date: | 2022-10-05 | | Last modified: | 2022-11-23 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Cryo-EM structures of light-harvesting 2 complexes from Rhodopseudomonas palustris reveal the molecular origin of absorption tuning.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7ZE3
 
 | | PucD-LH2 complex from Rps. palustris | | Descriptor: | (3'E)-3',4'-didehydro-1,2-dihydro-psi,psi-caroten-1-ol, BACTERIOCHLOROPHYLL A, Light-harvesting protein B-800-850 alpha chain, ... | | Authors: | Qian, P, Cogdell, R.J, Nguyen-Phan, T.C. | | Deposit date: | 2022-03-30 | | Release date: | 2022-10-05 | | Last modified: | 2022-11-23 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Cryo-EM structures of light-harvesting 2 complexes from Rhodopseudomonas palustris reveal the molecular origin of absorption tuning.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7ZE8
 
 | | PucE-LH2 complex from Rps. palustris | | Descriptor: | 1,2-Dihydro-psi,psi-caroten-1-ol, BACTERIOCHLOROPHYLL A, Light-harvesting protein, ... | | Authors: | Qian, P, Cogdell, R.J, Nguyen-Phan, T.C. | | Deposit date: | 2022-03-30 | | Release date: | 2022-10-05 | | Last modified: | 2022-11-23 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Cryo-EM structures of light-harvesting 2 complexes from Rhodopseudomonas palustris reveal the molecular origin of absorption tuning.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7ZCU
 
 | | PucA-LH2 complex from Rps. palustris | | Descriptor: | 1,2-Dihydro-psi,psi-caroten-1-ol, BACTERIOCHLOROPHYLL A, Light-harvesting protein B-800-850 alpha chain, ... | | Authors: | Qian, P, Cogdell, R.J. | | Deposit date: | 2022-03-28 | | Release date: | 2022-10-12 | | Last modified: | 2022-11-23 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Cryo-EM structures of light-harvesting 2 complexes from Rhodopseudomonas palustris reveal the molecular origin of absorption tuning.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
1O9B
 
 | | QUINATE/SHIKIMATE DEHYDROGENASE YDIB COMPLEXED WITH NADH | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, HYPOTHETICAL SHIKIMATE 5-DEHYDROGENASE-LIKE PROTEIN YDIB, PHOSPHATE ION | | Authors: | Michel, G, Cygler, M. | | Deposit date: | 2002-12-12 | | Release date: | 2003-02-01 | | Last modified: | 2013-10-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structures of Shikimate Dehydrogenase Aroe and its Paralog Ydib: A Common Structural Framework for Different Activities
J.Biol.Chem., 278, 2003
|
|
5V8K
 
 | | Homodimeric reaction center of H. modesticaldum | | Descriptor: | 1-[GLYCEROLYLPHOSPHONYL]-2-[8-(2-HEXYL-CYCLOPROPYL)-OCTANAL-1-YL]-3-[HEXADECANAL-1-YL]-GLYCEROL, 4,4'-Diaponeurosporene, 8(1)-OH-Chlorophyll aF, ... | | Authors: | Gisriel, C, Sarrou, I, Ferlez, B, Golbeck, J, Redding, K.E, Fromme, R. | | Deposit date: | 2017-03-22 | | Release date: | 2017-09-06 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of a symmetric photosynthetic reaction center-photosystem.
Science, 357, 2017
|
|
