4RS1
 
 | | Crystal structure of receptor-cytokine complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, Granulocyte-macrophage colony-stimulating factor, ... | | Authors: | Broughton, S.E, Parker, M.W, Dhagat, U. | | Deposit date: | 2014-11-06 | | Release date: | 2015-11-11 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.68 Å) | | Cite: | Conformational Changes in the GM-CSF Receptor Suggest a Molecular Mechanism for Affinity Conversion and Receptor Signaling.
Structure, 24, 2016
|
|
19GS
 
 | | Glutathione s-transferase p1-1 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 3,3'-(4,5,6,7-TETRABROMO-3-OXO-1(3H)-ISOBENZOFURANYLIDENE)BIS [6-HYDROXYBENZENESULFONIC ACID]ANION, GLUTATHIONE, ... | | Authors: | Oakley, A.J, Lo Bello, M, Parker, M.W. | | Deposit date: | 1997-12-14 | | Release date: | 1998-12-30 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The ligandin (non-substrate) binding site of human Pi class glutathione transferase is located in the electrophile binding site (H-site).
J.Mol.Biol., 291, 1999
|
|
1A87
 
 | | COLICIN N | | Descriptor: | COLICIN N | | Authors: | Vetter, I.R, Parker, M.W, Tucker, A.D, Lakey, J.H, Pattus, F, Tsernoglou, D. | | Deposit date: | 1998-04-03 | | Release date: | 1999-04-06 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Crystal structure of a colicin N fragment suggests a model for toxicity.
Structure, 6, 1998
|
|
6VQS
 
 | |
6VX2
 
 | |
6W0U
 
 | |
6W42
 
 | |
1FW1
 
 | | Glutathione transferase zeta/maleylacetoacetate isomerase | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, GLUTATHIONE, GLUTATHIONE TRANSFERASE ZETA, ... | | Authors: | Polekhina, G, Board, P.G, Blackburn, A.C, Parker, M.W. | | Deposit date: | 2000-09-20 | | Release date: | 2001-09-26 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of maleylacetoacetate isomerase/glutathione transferase zeta reveals the molecular basis for its remarkable catalytic promiscuity.
Biochemistry, 40, 2001
|
|
8V5T
 
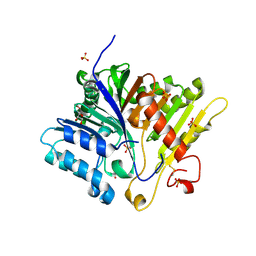 | | Crystal structure of Alzheimers disease phospholipase D3 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 5'-3' exonuclease PLD3, GLYCEROL, ... | | Authors: | Ishii, K, Hermans, S.J, Nero, T.L, Gorman, M.A, Parker, M.W. | | Deposit date: | 2023-12-01 | | Release date: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of Alzheimer's disease phospholipase D3 provides a molecular basis for understanding its normal and pathological functions.
Febs J., 2024
|
|
7SIA
 
 | |
6NAL
 
 | |
6NMY
 
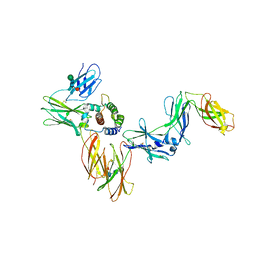 | | A Cytokine-receptor complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Cytokine receptor common subunit beta, ... | | Authors: | Dhagat, U, Kan, W.L, Hercus, T.R, Broughton, S.E, Nero, T.L, Lopez, A.F, Parker, M.W. | | Deposit date: | 2019-01-13 | | Release date: | 2020-01-22 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.301 Å) | | Cite: | Distinct Assemblies of Heterodimeric Cytokine Receptors Govern Stemness Programs in Leukemia.
Cancer Discov, 13, 2023
|
|
4Y0G
 
 | | beta2 carbohydrate binding module (CBM) of AMP-activated protein kinase (AMPK) | | Descriptor: | 5'-AMP-activated protein kinase subunit beta-2, GLYCEROL | | Authors: | Mobbs, J, Gorman, M.A, Parker, M.W, Gooley, P.R, Griffin, M. | | Deposit date: | 2015-02-06 | | Release date: | 2015-04-08 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Determinants of oligosaccharide specificity of the carbohydrate-binding modules of AMP-activated protein kinase.
Biochem.J., 468, 2015
|
|
6OIR
 
 | | Crystal structure of MYST acetyltransferase domain in complex with inhibitor 62 | | Descriptor: | 4-fluoro-N'-(phenylsulfonyl)[1,1'-biphenyl]-3-carbohydrazide, GLYCEROL, Histone acetyltransferase KAT8, ... | | Authors: | Hermans, S.J, Chung, M.C, Parker, M.W, Thomas, T, Baell, J.B. | | Deposit date: | 2019-04-09 | | Release date: | 2019-07-03 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Discovery of Benzoylsulfonohydrazides as Potent Inhibitors of the Histone Acetyltransferase KAT6A.
J.Med.Chem., 62, 2019
|
|
7TBA
 
 | | Pentraxin - ligand complex | | Descriptor: | C-reactive protein, CALCIUM ION, [3-(dibutylamino)propyl]phosphonic acid | | Authors: | Shing, K.S.C.T, Morton, C.J, Parker, M.W. | | Deposit date: | 2021-12-21 | | Release date: | 2022-10-19 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | A novel phosphocholine-mimetic inhibits a pro-inflammatory conformational change in C-reactive protein.
Embo Mol Med, 15, 2023
|
|
7L1P
 
 | | HIV Integrase Core domain (IN) in complex with dimer-spanning ligand | | Descriptor: | (2-{[3-(4-{2-[(3-{[3-(carboxymethyl)-5-methyl-1-benzofuran-2-yl]ethynyl}benzene-1-carbonyl)amino]ethyl}piperazine-1-carbonyl)phenyl]ethynyl}-5-methyl-1-benzofuran-3-yl)acetic acid, IODIDE ION, Integrase, ... | | Authors: | Gorman, M.A, Parker, M.W. | | Deposit date: | 2020-12-15 | | Release date: | 2021-12-22 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | HIV Integrase core domain in complex with inhibitor
To Be Published
|
|
7L2Y
 
 | |
6OWI
 
 | | Crystal structure of MYST acetyltransferase domain in complex with inhibitor 85 | | Descriptor: | GLYCEROL, Histone acetyltransferase KAT8, N'-[(2-fluorophenyl)sulfonyl]benzohydrazide, ... | | Authors: | Hermans, S.J, Chung, M.C, Parker, M.W, Thomas, T, Baell, J.B. | | Deposit date: | 2019-05-09 | | Release date: | 2020-04-01 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Discovery of Acylsulfonohydrazide-Derived Inhibitors of the Lysine Acetyltransferase, KAT6A, as Potent Senescence-Inducing Anti-Cancer Agents.
J.Med.Chem., 63, 2020
|
|
6OIN
 
 | | Crystal structure of MYST acetyltransferase domain in complex with inhibitor CTX-124143 | | Descriptor: | 2-fluoro-N'-[(naphthalen-2-yl)sulfonyl]benzohydrazide, GLYCEROL, Histone acetyltransferase KAT8, ... | | Authors: | Hermans, S.J, Chung, M.C, Parker, M.W, Thomas, T, Baell, J.B. | | Deposit date: | 2019-04-09 | | Release date: | 2019-07-03 | | Last modified: | 2019-08-21 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Discovery of Benzoylsulfonohydrazides as Potent Inhibitors of the Histone Acetyltransferase KAT6A.
J.Med.Chem., 62, 2019
|
|
7LO4
 
 | |
2AN6
 
 | | Protein-peptide complex | | Descriptor: | Ubiquitin ligase SIAH1A, ZINC ION, peptide from Phyllopod | | Authors: | House, C.M, Hancock, N.C, Moller, A, Cromer, B.A, Fedorov, V, Bowtell, D.D.L, Parker, M.W, Polekhina, G. | | Deposit date: | 2005-08-11 | | Release date: | 2006-05-30 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Elucidation of the substrate binding site of Siah ubiquitin ligase
Structure, 14, 2006
|
|
2FK3
 
 | |
2FJZ
 
 | |
2FKL
 
 | |
2FK1
 
 | |
