1X0T
 
 | | Crystal structure of ribonuclease P protein Ph1601p from Pyrococcus horikoshii OT3 | | Descriptor: | Ribonuclease P protein component 4, ZINC ION | | Authors: | Kakuta, Y, Ishimatsu, I, Numata, T, Kimura, K, Yao, M, Tanaka, I, Kimura, M. | | Deposit date: | 2005-03-29 | | Release date: | 2005-11-15 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal Structure of a Ribonuclease P Protein Ph1601p from Pyrococcus horikoshii OT3: An Archaeal Homologue of Human Nuclear Ribonuclease P Protein Rpp21(,)
Biochemistry, 44, 2005
|
|
6JMS
 
 | | CJP38, a beta-1,3-glucanase and allergen of Cryptomeria japonica pollen | | Descriptor: | 1,2-ETHANEDIOL, Pollen allergen CJP38 | | Authors: | Takashima, T, Numata, T, Fukamizo, T, Ohnuma, T. | | Deposit date: | 2019-03-13 | | Release date: | 2020-05-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | CJP38, a beta-1,3-glucanase and allergen of Cryptomeria japonica pollen
To Be Published
|
|
5BUM
 
 | | Crystal Structure of LysM domain from Equisetum arvense chitinase A | | Descriptor: | Chitinase A, SULFATE ION | | Authors: | Kitaoku, Y, Numata, T, Ohnuma, T, Taira, T, Fukamizo, T. | | Deposit date: | 2015-06-04 | | Release date: | 2016-06-08 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure, mechanism, and phylogeny of LysM-chitinase conjugates specifically found in fern plants.
Plant Sci., 321, 2022
|
|
3X1L
 
 | |
4ZXE
 
 | | X-ray crystal structure of chitosan-binding module 1 derived from chitosanase/glucanase from Paenibacillus sp. IK-5. | | Descriptor: | 1,2-ETHANEDIOL, Glucanase/Chitosanase, SULFATE ION | | Authors: | Shinya, S, Oi, H, Kitaoku, Y, Ohnuma, T, Numata, T, Fukamizo, T. | | Deposit date: | 2015-05-20 | | Release date: | 2016-04-13 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Mechanism of chitosan recognition by CBM32 carbohydrate-binding modules from a Paenibacillus sp. IK-5 chitosanase/glucanase
Biochem.J., 473, 2016
|
|
4ZY9
 
 | | X-ray crystal structure of selenomethionine-labelled V110M mutant of chitosan-binding module 1 derived from chitosanase/glucanase from Paenibacillus sp. IK-5 | | Descriptor: | Glucanase/chitosanase | | Authors: | Shinya, S, Oi, H, Kitaoku, Y, Ohnuma, T, Numata, T, Fukamizo, T. | | Deposit date: | 2015-05-21 | | Release date: | 2016-04-13 | | Last modified: | 2020-02-19 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Mechanism of chitosan recognition by CBM32 carbohydrate-binding modules from a Paenibacillus sp. IK-5 chitosanase/glucanase
Biochem.J., 473, 2016
|
|
4ZZ5
 
 | | X-ray crystal structure of chitosan-binding module 2 derived from chitosanase/glucanase from Paenibacillus sp. IK-5 | | Descriptor: | 1,2-ETHANEDIOL, Glucanase/chitosanase, SULFATE ION | | Authors: | Shinya, S, Oi, H, Kitaoku, Y, Ohnuma, T, Numata, T, Fukamizo, T. | | Deposit date: | 2015-05-22 | | Release date: | 2016-04-13 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.29 Å) | | Cite: | Mechanism of chitosan recognition by CBM32 carbohydrate-binding modules from a Paenibacillus sp. IK-5 chitosanase/glucanase
Biochem.J., 473, 2016
|
|
4ZZ8
 
 | | X-ray crystal structure of chitosan-binding module 2 in complex with chitotriose derived from chitosanase/glucanase from Paenibacillus sp. IK-5 | | Descriptor: | 1,2-ETHANEDIOL, 2-amino-2-deoxy-beta-D-glucopyranose-(1-4)-2-amino-2-deoxy-beta-D-glucopyranose-(1-4)-2-amino-2-deoxy-beta-D-glucopyranose, Glucanase/chitosanase, ... | | Authors: | Shinya, S, Oi, H, Kitaoku, Y, Ohnuma, T, Numata, T, Fukamizo, T. | | Deposit date: | 2015-05-22 | | Release date: | 2016-04-13 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Mechanism of chitosan recognition by CBM32 carbohydrate-binding modules from a Paenibacillus sp. IK-5 chitosanase/glucanase
Biochem.J., 473, 2016
|
|
1UAX
 
 | |
2CZW
 
 | | Crystal structure analysis of protein component Ph1496p of P.horikoshii ribonuclease P | | Descriptor: | 50S ribosomal protein L7Ae | | Authors: | Fukuhara, H, Kifusa, M, Watanabe, M, Terada, A, Honda, T, Numata, T, Kakuta, Y, Kimura, M. | | Deposit date: | 2005-07-19 | | Release date: | 2006-04-25 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A fifth protein subunit Ph1496p elevates the optimum temperature for the ribonuclease P activity from Pyrococcus horikoshii OT3
Biochem.Biophys.Res.Commun., 343, 2006
|
|
1UCD
 
 | | Crystal structure of Ribonuclease MC1 from bitter gourd seeds complexed with 5'-UMP | | Descriptor: | Ribonuclease MC, URACIL, URIDINE-5'-MONOPHOSPHATE | | Authors: | Suzuki, A, Numata, T, Yao, M, Kimura, M, Tanaka, I. | | Deposit date: | 2003-04-10 | | Release date: | 2004-05-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structure of RNase MC1 from bitter gourd seeds in complex with 5'UMP
To be published
|
|
1V9H
 
 | | Crystal structure of the RNase MC1 mutant Y101A in complex with 5'-UMP | | Descriptor: | Ribonuclease MC, SULFATE ION, URIDINE-5'-MONOPHOSPHATE | | Authors: | Kimura, K, Numata, T, Kakuta, Y, Kimura, M. | | Deposit date: | 2004-01-26 | | Release date: | 2004-10-05 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Amino acids conserved at the C-terminal half of the ribonuclease t2 family contribute to protein stability of the enzymes
Biosci.Biotechnol.Biochem., 68, 2004
|
|
1V77
 
 | | Crystal structure of the PH1877 protein | | Descriptor: | hypothetical protein PH1877 | | Authors: | Takagi, H, Numata, T, Kakuta, Y, Kimura, M. | | Deposit date: | 2003-12-12 | | Release date: | 2004-08-31 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of the ribonuclease P protein Ph1877p from hyperthermophilic archaeon Pyrococcus horikoshii OT3
Biochem.Biophys.Res.Commun., 319, 2004
|
|
1UCA
 
 | | Crystal structure of the Ribonuclease MC1 from bitter gourd seeds complexed with 2'-UMP | | Descriptor: | PHOSPHORIC ACID MONO-[2-(2,4-DIOXO-3,4-DIHYDRO-2H-PYRIMIDIN-1-YL)-4-HYDROXY-5-HYDROXYMETHYL-TETRAHYDRO-FURAN-3-YL] ESTER, Ribonuclease MC | | Authors: | Suzuki, A, Yao, M, Tanaka, I, Numata, T, Kikukawa, S, Yamasaki, N, Kimura, M. | | Deposit date: | 2003-04-10 | | Release date: | 2003-04-29 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Crystal structures of the ribonuclease MC1 from bitter gourd seeds, complexed with 2'-UMP or 3'-UMP, reveal structural basis for uridine specificity
Biochem.Biophys.Res.Commun., 275, 2000
|
|
1UCC
 
 | | Crystal structure of the Ribonuclease MC1 from bitter gourd seeds complexed with 3'-UMP. | | Descriptor: | 3'-URIDINEMONOPHOSPHATE, Ribonuclease MC | | Authors: | Suzuki, A, Yao, M, Tanaka, I, Numata, T, Kikukawa, S, Yamasaki, N, Kimura, M. | | Deposit date: | 2003-04-10 | | Release date: | 2003-04-29 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Crystal structures of the ribonuclease MC1 from bitter gourd seeds, complexed with 2'-UMP or 3'-UMP, reveal structural basis for uridine specificity
Biochem.Biophys.Res.Commun., 275, 2000
|
|
7XMH
 
 | | Crystal structure of a rice class IIIb chitinase, Oschib2 | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, Putative class III chitinase | | Authors: | Jun, T, Tomoya, T, Tomoyuki, N, Takayuki, O. | | Deposit date: | 2022-04-25 | | Release date: | 2023-05-03 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.18 Å) | | Cite: | Characterization of two rice GH18 chitinases belonging to family 8 of plant pathogenesis-related proteins.
Plant Sci., 326, 2023
|
|
6M04
 
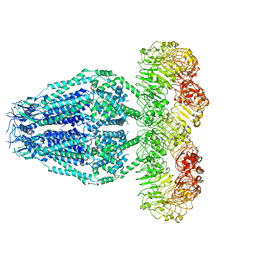 | | Structure of the human homo-hexameric LRRC8D channel at 4.36 Angstroms | | Descriptor: | Volume-regulated anion channel subunit LRRC8D | | Authors: | Nakamura, R, Kasuya, G, Yokoyama, T, Shirouzu, M, Ishitani, R, Nureki, O. | | Deposit date: | 2020-02-20 | | Release date: | 2020-06-17 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (4.36 Å) | | Cite: | Cryo-EM structure of the volume-regulated anion channel LRRC8D isoform identifies features important for substrate permeation.
Commun Biol, 3, 2020
|
|
2RVA
 
 | |
2RV9
 
 | |
3H39
 
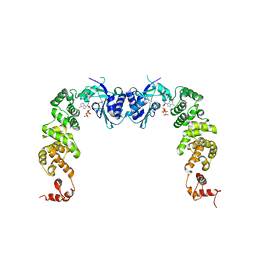 | | The complex structure of CCA-adding enzyme with ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, TRNA nucleotidyl transferase-related protein | | Authors: | Toh, Y, Tomita, K. | | Deposit date: | 2009-04-16 | | Release date: | 2009-10-13 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.854 Å) | | Cite: | Mechanism for the definition of elongation and termination by the class II CCA-adding enzyme
Embo J., 28, 2009
|
|
3H38
 
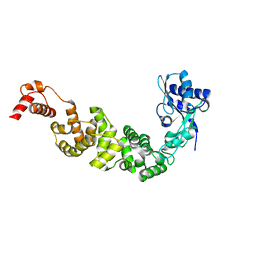 | | The structure of CCA-adding enzyme apo form II | | Descriptor: | TRNA nucleotidyl transferase-related protein | | Authors: | Toh, Y, Tomita, K. | | Deposit date: | 2009-04-16 | | Release date: | 2009-10-13 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | Mechanism for the definition of elongation and termination by the class II CCA-adding enzyme
Embo J., 28, 2009
|
|
3H37
 
 | | The structure of CCA-adding enzyme apo form I | | Descriptor: | TRNA nucleotidyl transferase-related protein | | Authors: | Toh, Y, Tomita, K. | | Deposit date: | 2009-04-16 | | Release date: | 2009-10-13 | | Last modified: | 2014-02-05 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Mechanism for the definition of elongation and termination by the class II CCA-adding enzyme
Embo J., 28, 2009
|
|
7E15
 
 | |
3H3A
 
 | | The complex structure of CCA-adding enzyme with CTP | | Descriptor: | CYTIDINE-5'-TRIPHOSPHATE, TRNA nucleotidyl transferase-related protein | | Authors: | Toh, Y, Tomita, K. | | Deposit date: | 2009-04-16 | | Release date: | 2009-10-13 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.801 Å) | | Cite: | Mechanism for the definition of elongation and termination by the class II CCA-adding enzyme
Embo J., 28, 2009
|
|
2D6F
 
 | |
