6HFY
 
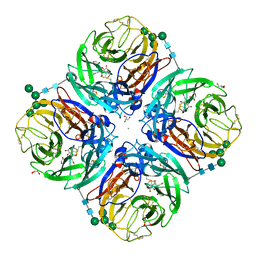 | | Influenza A virus N6 neuraminidase complex with DANA (Duck/England/56). | | Descriptor: | 2-DEOXY-2,3-DEHYDRO-N-ACETYL-NEURAMINIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Salinger, M.T, Hobbs, J.R, Murray, J.W, Laver, W.G, Kuhn, P, Garman, E.F. | | Deposit date: | 2018-08-22 | | Release date: | 2018-08-29 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | High Resolution Structures of Viral Neuraminidase with Drugs Bound in the Active Site. (In preparation)
To Be Published
|
|
6SDM
 
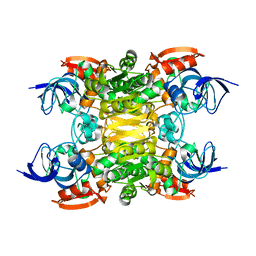 | | NADH-dependent variant of TBADH | | Descriptor: | NADP-dependent isopropanol dehydrogenase, ZINC ION | | Authors: | Selles Vidal, L, Murray, J.W, Heap, J.T. | | Deposit date: | 2019-07-28 | | Release date: | 2020-08-26 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Versatile selective evolutionary pressure using synthetic defect in universal metabolism.
Nat Commun, 12, 2021
|
|
6SCH
 
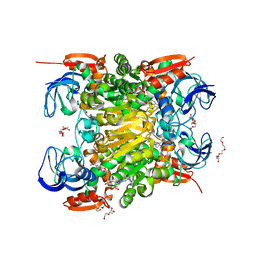 | | NADH-dependent variant of CBADH | | Descriptor: | MAGNESIUM ION, NADP-dependent isopropanol dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Selles Vidal, L, Murray, J.W, Heap, J.T. | | Deposit date: | 2019-07-24 | | Release date: | 2020-08-26 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Versatile selective evolutionary pressure using synthetic defect in universal metabolism.
Nat Commun, 12, 2021
|
|
8ASL
 
 | | RCII/PSI complex, class 2 | | Descriptor: | (1R,2S)-4-{(1E,3E,5E,7E,9E,11E,13E,15E,17E)-18-[(4S)-4-hydroxy-2,6,6-trimethylcyclohex-1-en-1-yl]-3,7,12,16-tetramethyloctadeca-1,3,5,7,9,11,13,15,17-nonaen-1-yl}-2,5,5-trimethylcyclohex-3-en-1-ol, (3'R)-3'-hydroxy-beta,beta-caroten-4-one, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, ... | | Authors: | Zhao, Z, Vercellino, I, Knoppova, J, Sobotka, R, Murray, J.W, Nixon, P.J, Sazanov, L.A, Komenda, J. | | Deposit date: | 2022-08-19 | | Release date: | 2023-08-16 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.15 Å) | | Cite: | The Ycf48 accessory factor occupies the site of the oxygen-evolving manganese cluster during photosystem II biogenesis.
Nat Commun, 14, 2023
|
|
8AM5
 
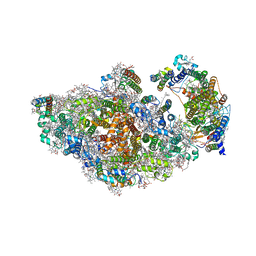 | | RCII/PSI complex, class 3 | | Descriptor: | (1R,2S)-4-{(1E,3E,5E,7E,9E,11E,13E,15E,17E)-18-[(4S)-4-hydroxy-2,6,6-trimethylcyclohex-1-en-1-yl]-3,7,12,16-tetramethyloctadeca-1,3,5,7,9,11,13,15,17-nonaen-1-yl}-2,5,5-trimethylcyclohex-3-en-1-ol, (3'R)-3'-hydroxy-beta,beta-caroten-4-one, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, ... | | Authors: | Zhao, Z, Vercellino, I, Knoppova, J, Sobotka, R, Murray, J.W, Nixon, P.J, Sazanov, L.A, Komenda, J. | | Deposit date: | 2022-08-02 | | Release date: | 2023-08-16 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | The Ycf48 accessory factor occupies the site of the oxygen-evolving manganese cluster during photosystem II biogenesis.
Nat Commun, 14, 2023
|
|
8ASP
 
 | | RCII/PSI complex, focused refinement of PSI | | Descriptor: | (1R,2S)-4-{(1E,3E,5E,7E,9E,11E,13E,15E,17E)-18-[(4S)-4-hydroxy-2,6,6-trimethylcyclohex-1-en-1-yl]-3,7,12,16-tetramethyloctadeca-1,3,5,7,9,11,13,15,17-nonaen-1-yl}-2,5,5-trimethylcyclohex-3-en-1-ol, (3'R)-3'-hydroxy-beta,beta-caroten-4-one, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, ... | | Authors: | Zhao, Z, Vercellino, I, Knoppova, J, Sobotka, R, Murray, J.W, Nixon, P.J, Sazanov, L.A, Komenda, J. | | Deposit date: | 2022-08-20 | | Release date: | 2023-08-16 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | The Ycf48 accessory factor occupies the site of the oxygen-evolving manganese cluster during photosystem II biogenesis.
Nat Commun, 14, 2023
|
|
4ZGS
 
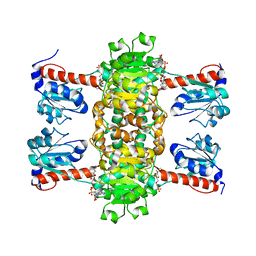 | | Identification of the pyruvate reductase of Chlamydomonas reinhardtii | | Descriptor: | NICOTINAMIDE-ADENINE-DINUCLEOTIDE, Putative D-lactate dehydrogenase | | Authors: | Burgess, S.J, Hussein, T, Yeoman, J.A, Iamshanova, O, Boehm, M, Bundy, J, Bialek, W, Murray, J.W, Nixon, P.J. | | Deposit date: | 2015-04-23 | | Release date: | 2015-12-02 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.461 Å) | | Cite: | Identification of the Elusive Pyruvate Reductase of Chlamydomonas reinhardtii Chloroplasts.
Plant Cell.Physiol., 57, 2016
|
|
5A5L
 
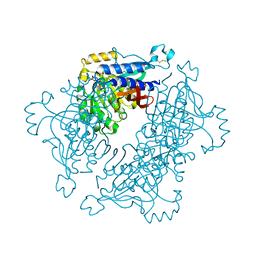 | | Structure of dual function FBPase SBPase from Thermosynechococcus elongatus | | Descriptor: | 7-O-phosphono-alpha-L-galacto-hept-2-ulopyranose, D-FRUCTOSE 1,6-BISPHOSPHATASE CLASS 2/SEDOHEPTULOSE 1,7-BISPHOSPHATASE, MAGNESIUM ION, ... | | Authors: | Cotton, C.A.R, Kabasakal, B, Miah, N, Murray, J.W. | | Deposit date: | 2015-06-19 | | Release date: | 2015-10-14 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Structure of the Dual-Function Fructose-1,6/Sedoheptulose-1, 7-Bisphosphatase from Thermosynechococcus Elongatus Bound with Sedoheptulose-7-Phosphate.
Acta Crystallogr.,Sect.F, 71, 2015
|
|
6GHL
 
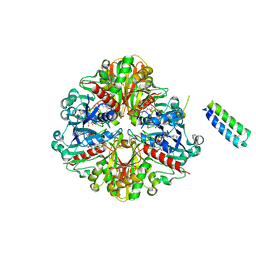 | | cyanobacterial GAPDH with full-length CP12 | | Descriptor: | CP12 polypeptide, Glyceraldehyde-3-phosphate dehydrogenase, MALONATE ION, ... | | Authors: | McFarlane, C.R, Murray, J.W. | | Deposit date: | 2018-05-08 | | Release date: | 2019-05-08 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.378 Å) | | Cite: | Structural basis of light-induced redox regulation in the Calvin-Benson cycle in cyanobacteria.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
5ABR
 
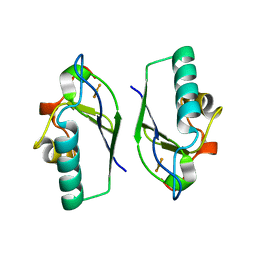 | |
2Y6X
 
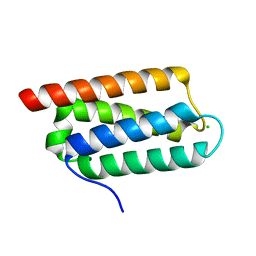 | | Structure of Psb27 from Thermosynechococcus elongatus | | Descriptor: | CHLORIDE ION, PHOTOSYSTEM II 11 KD PROTEIN | | Authors: | Michoux, F, Takasaka, K, Boehm, M, Nixon, P.J, Murray, J.W. | | Deposit date: | 2011-01-27 | | Release date: | 2012-01-11 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal Structure of the Psb27 Assembly Factor at 1.6A: Implications for Binding to Photosystem II.
Photosynth.Res., 110, 2012
|
|
5DI5
 
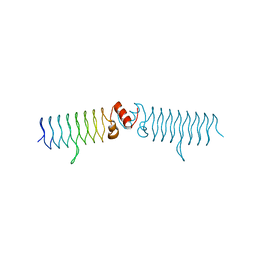 | | beta1 t801 loop variant in P3221 | | Descriptor: | beta1 t801 loop variant | | Authors: | MacDonald, J.T, Kabasakal, B.V, Murray, J.W. | | Deposit date: | 2015-08-31 | | Release date: | 2016-08-31 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Synthetic beta-solenoid proteins with the fragment-free computational design of a beta-hairpin extension.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5DNS
 
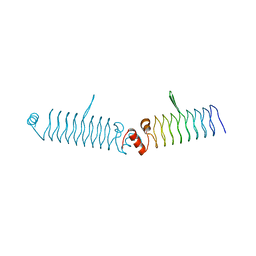 | | t1428 loop variant in P3221 | | Descriptor: | t1428 beta1 loop design | | Authors: | MacDonald, J.T, Kabasakal, B.V, Murray, J.W. | | Deposit date: | 2015-09-10 | | Release date: | 2016-09-28 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.561 Å) | | Cite: | Synthetic beta-solenoid proteins with the fragment-free computational design of a beta-hairpin extension.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
2J6Z
 
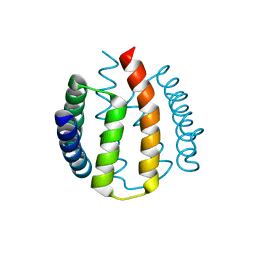 | | Structural and functional characterisation of partner-switching regulating the environmental stress response in B. subtilis | | Descriptor: | PHOSPHOSERINE PHOSPHATASE RSBU | | Authors: | Hardwick, S.W, Pane-Farre, J, Delumeau, O, Marles-Wright, J, Murray, J.W, Hecker, M, Lewis, R.J. | | Deposit date: | 2006-10-05 | | Release date: | 2007-02-13 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural and functional characterization of partner switching regulating the environmental stress response in Bacillus subtilis.
J. Biol. Chem., 282, 2007
|
|
2J70
 
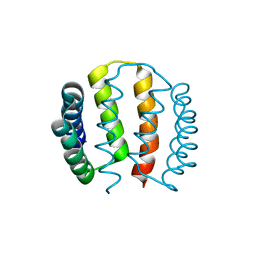 | | Structural and functional characterisation of partner-switching regulating the environmental stress response in B. subtilis | | Descriptor: | PHOSPHOSERINE PHOSPHATASE RSBU | | Authors: | Hardwick, S.W, Pane-Farre, J, Delumeau, O, Marles-Wright, J, Murray, J.W, Hecker, M, Lewis, R.J. | | Deposit date: | 2006-10-05 | | Release date: | 2007-02-13 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural and functional characterization of partner switching regulating the environmental stress response in Bacillus subtilis.
J. Biol. Chem., 282, 2007
|
|
5FRT
 
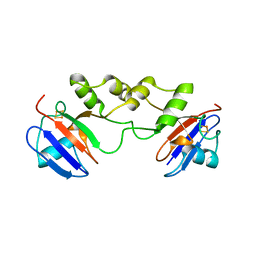 | |
2J6Y
 
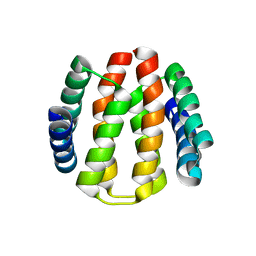 | | Structural and Functional Characterisation of partner switching regulating the environmental stress response in Bacillus subtilis | | Descriptor: | PHOSPHOSERINE PHOSPHATASE RSBU | | Authors: | Hardwick, S.W, Pane-Farre, J, Delumeau, O, Marles-Wright, J, Murray, J.W, Hecker, M, Lewis, R.J. | | Deposit date: | 2006-10-05 | | Release date: | 2007-02-13 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural and functional characterization of partner switching regulating the environmental stress response in Bacillus subtilis.
J. Biol. Chem., 282, 2007
|
|
5DN0
 
 | | t1555 loop variant | | Descriptor: | beta1 t1555 loop variant | | Authors: | MacDonald, J.T, Kabasakal, B.V, Murray, J.W. | | Deposit date: | 2015-09-09 | | Release date: | 2016-08-31 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (4.4 Å) | | Cite: | Synthetic beta-solenoid proteins with the fragment-free computational design of a beta-hairpin extension.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5LYP
 
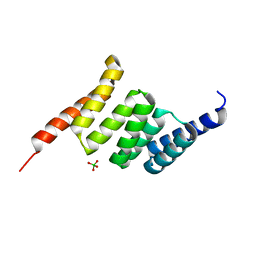 | | Crystal structure of the Tpr Domain of Sgt2. | | Descriptor: | BORATE ION, Small glutamine-rich tetratricopeptide repeat-containing protein 2 | | Authors: | Krysztofinska, E.M, Morgan, R.M.L, Thapaliya, A, Evans, N.J, Murray, J.W, Martinez-Lumbreras, S, Isaacson, R.L. | | Deposit date: | 2016-09-28 | | Release date: | 2017-10-25 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structure and Interactions of the TPR Domain of Sgt2 with Yeast Chaperones and Ybr137wp.
Front Mol Biosci, 4, 2017
|
|
5LYN
 
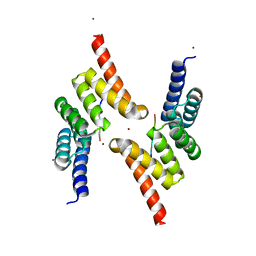 | | Structure of the Tpr Domain of Sgt2 in complex with yeast Ssa1 peptide fragment | | Descriptor: | PRO-THR-VAL-GLU-GLU-VAL-ASP, Small glutamine-rich tetratricopeptide repeat-containing protein 2, ZINC ION | | Authors: | Krysztofinska, E.M, Evans, N.J, Thapaliya, A, Murray, J.W, Morgan, R.M.L, Martinez-Lumbreras, S, Isaacson, R.L. | | Deposit date: | 2016-09-28 | | Release date: | 2017-10-25 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure and Interactions of the TPR Domain of Sgt2 with Yeast Chaperones and Ybr137wp.
Front Mol Biosci, 4, 2017
|
|
5SVB
 
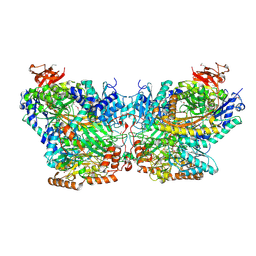 | | Mechanism of ATP-Dependent Acetone Carboxylation, Acetone Carboxylase AMP bound structure | | Descriptor: | ADENOSINE MONOPHOSPHATE, Acetone carboxylase alpha subunit, Acetone carboxylase beta subunit, ... | | Authors: | Eilers, B.J, Mus, F, Alleman, A.B, Kabasakal, B.V, Murray, J.W, Nocek, B.P, Dubois, J.L, Peters, J.W. | | Deposit date: | 2016-08-05 | | Release date: | 2017-08-09 | | Last modified: | 2019-12-04 | | Method: | X-RAY DIFFRACTION (2.645 Å) | | Cite: | Structural Basis for the Mechanism of ATP-Dependent Acetone Carboxylation.
Sci Rep, 7, 2017
|
|
5SVC
 
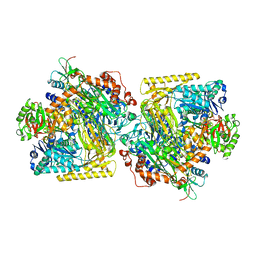 | | Mechanism of ATP-Dependent Acetone Carboxylation, Acetone Carboxylase nucleotide-free structure | | Descriptor: | Acetone carboxylase alpha subunit, Acetone carboxylase beta subunit, Acetone carboxylase gamma subunit, ... | | Authors: | Eilers, B.J, Mus, F, Alleman, A.B, Kabasakal, B.V, Murray, J.W, Nocek, B.P, Dubois, J.L, Peters, J.W. | | Deposit date: | 2016-08-05 | | Release date: | 2017-08-09 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural Basis for the Mechanism of ATP-Dependent Acetone Carboxylation.
Sci Rep, 7, 2017
|
|
3EPY
 
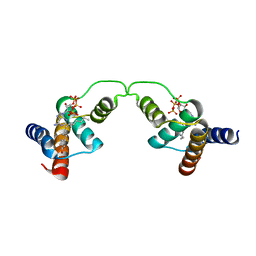 | | Crystal Structure of human acyl-CoA binding domain 7 complexed with palmitoyl-Coa | | Descriptor: | Acyl-CoA-binding domain-containing protein 7, COENZYME A, PALMITIC ACID | | Authors: | Kavanagh, K.L, Salah, E, Yue, W.W, Savitsky, P, Murray, J.W, Arrowsmith, C.H, Weigelt, J, Edwards, A.M, Bountra, C, von Delft, F, Oppermann, U, Structural Genomics Consortium (SGC) | | Deposit date: | 2008-09-30 | | Release date: | 2009-01-20 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.005 Å) | | Cite: | Crystal Structure of human acyl-CoA binding domain 7 complexed with palmitoyl-Coa
To be Published
|
|
3F2N
 
 | | Crystal Structure of Human Haspin with an Imidazo-pyridazine ligand | | Descriptor: | (2S)-2-{[3-(3-aminophenyl)imidazo[1,2-b]pyridazin-6-yl]amino}-3-methylbutan-1-ol, 1,2-ETHANEDIOL, PHOSPHATE ION, ... | | Authors: | Filippakopoulos, P, Eswaran, J, Keates, T, Burgess-Brown, N, Fedorov, O, Yue, W.W, Murray, J.W, Pike, A.C.W, Von Delft, F, Arrowsmith, C.H, Edwards, A.M, Weigelt, J, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2008-10-30 | | Release date: | 2008-12-02 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of Human Haspin with an Imidazo-pyridazine ligand
To be Published
|
|
3DLZ
 
 | | Crystal structure of human haspin in complex with AMP | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE MONOPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Filippakopoulos, P, Eswaran, J, Keates, T, Burgess-Brown, N, Murray, J.W, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Wickstroem, M, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2008-06-30 | | Release date: | 2008-07-15 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure and functional characterization of the atypical human kinase haspin.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
