3QJI
 
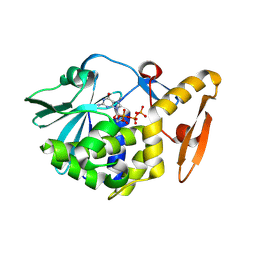 | | Crystal structure of the complex of ribosome inactivating protein with 7-methylguanosine triphosphate at 1.75A resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 7-METHYL-GUANOSINE-5'-TRIPHOSPHATE, Ribosome inactivating protein | | Authors: | Kumar, M, Kushwaha, G.S, Sinha, M, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2011-01-29 | | Release date: | 2011-03-09 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal structure of the complex of ribosome inactivating protein with 7-methylguanosine triphosphate at 1.75A resolution
TO BE PUBLISHED
|
|
3KRQ
 
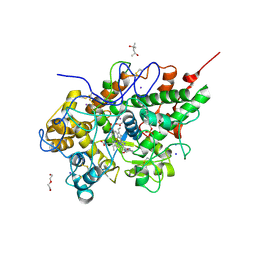 | | Crystal structure of the complex of lactoperoxidase with a potent inhibitor amino-triazole at 2.2a resolution | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, 2-acetamido-2-deoxy-alpha-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 3-AMINO-1,2,4-TRIAZOLE, ... | | Authors: | Singh, A.K, Singh, N, Sinha, M, Kushwaha, G.S, Kaur, P, Srinivasan, A, Sharma, S, Singh, T.P. | | Deposit date: | 2009-11-19 | | Release date: | 2010-05-26 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | First structural evidence for the mode of diffusion of aromatic ligands and ligand-induced closure of the hydrophobic channel in heme peroxidases
J.Biol.Inorg.Chem., 15, 2010
|
|
3NJS
 
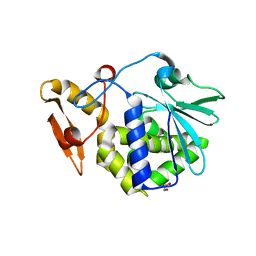 | | Crystal structure of the complex formed between typeI ribosome inactivating protein and lactose at 2.1A resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, Ribosome inactivating protein, ... | | Authors: | Pandey, N, Kushwaha, G.S, Sinha, M, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2010-06-18 | | Release date: | 2010-07-14 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of the complex formed between typeI ribosome inactivating protein and lactose at 2.1A resolution
To be Published
|
|
3N5D
 
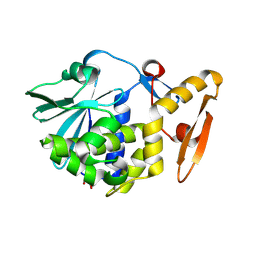 | | Crystal structure of the complex of type I ribosome inactivating protein with glucose at 1.9A resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, Ribosome inactivating protein, ... | | Authors: | Pandey, N, Kushwaha, G.S, Sinha, M, Kaur, P, Betzel, C, Sharma, S, Singh, T.P. | | Deposit date: | 2010-05-25 | | Release date: | 2010-06-30 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of the complex of type I ribosome inactivating protein with glucose at 1.9A resolution
To be Published
|
|
3N1D
 
 | | Crystal structure of the complex of type I ribosome inactivating protein with ribose at 1.7A resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Pandey, N, Kushwaha, G.S, Sinha, M, Kaur, P, Betzel, C, Sharma, S, Singh, T.P. | | Deposit date: | 2010-05-15 | | Release date: | 2010-06-23 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of the complex of type I ribosome inactivating protein with ribose at 1.7A resolution
To be Published
|
|
5GHU
 
 | | Crystal structure of YadV chaperone at 1.63 Angstrom | | Descriptor: | ACETATE ION, CHLORIDE ION, FORMIC ACID, ... | | Authors: | Pandey, N.K, Bhavesh, N.S. | | Deposit date: | 2016-06-20 | | Release date: | 2017-06-28 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Crystal structure of the usher chaperone YadV reveals a monomer with the proline lock in closed conformation suggestive of an intermediate state.
Febs Lett., 2020
|
|
2MY7
 
 | |
2MY8
 
 | |
