2YWW
 
 | | Crystal structure of aspartate carbamoyltransferase regulatory chain from Methanocaldococcus jannaschii | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Aspartate carbamoyltransferase regulatory chain, ZINC ION | | Authors: | Kanagawa, M, Baba, S, Kuramitsu, S, Yokoyama, S, Kawai, G, Sampei, G, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-04-23 | | Release date: | 2007-10-23 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of aspartate carbamoyltransferase regulatory chain from Methanocaldococcus jannaschii
To be Published
|
|
2YZJ
 
 | | Crystal structure of dCTP deaminase from Sulfolobus tokodaii | | Descriptor: | 167aa long hypothetical dUTPase, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, DEOXYURIDINE-5'-DIPHOSPHATE, ... | | Authors: | Kanagawa, M, Baba, S, Kuramitsu, S, Yokoyama, S, Kawai, G, Sampei, G, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-05-06 | | Release date: | 2007-11-06 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Crystal structure of dCTP deaminase from Sulfolobus tokodaii
To be Published
|
|
2ZDC
 
 | | Crystal structure of dUTPase from Sulfolobus tokodaii | | Descriptor: | 167aa long hypothetical dUTPase, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, SULFATE ION | | Authors: | Kanagawa, M, Baba S, Kuramitsu, S, Yokoyama, S, Kawai, G, Sampei, G, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-11-21 | | Release date: | 2008-05-27 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of dUTPase from Sulfolobus tokodaii
To be Published
|
|
3AQY
 
 | | Crystal structure of Plodia interpunctella beta-GRP/GNBP3 N-terminal domain | | Descriptor: | Beta-1,3-glucan-binding protein | | Authors: | Kanagawa, M, Satoh, T, Ikeda, A, Adachi, Y, Ohno, N, Yamaguchi, Y. | | Deposit date: | 2010-11-22 | | Release date: | 2011-06-22 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Structural insights into recognition of triple-helical beta-glucans by an insect fungal receptor
J.Biol.Chem., 286, 2011
|
|
3AQZ
 
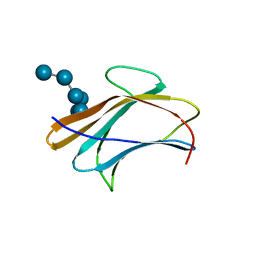 | | Crystal structure of Plodia interpunctella beta-GRP/GNBP3 N-terminal domain with laminarihexaoses | | Descriptor: | Beta-1,3-glucan-binding protein, beta-D-glucopyranose-(1-3)-beta-D-glucopyranose-(1-3)-beta-D-glucopyranose-(1-3)-beta-D-glucopyranose-(1-3)-beta-D-glucopyranose-(1-3)-beta-D-glucopyranose | | Authors: | Kanagawa, M, Satoh, T, Ikeda, A, Adachi, Y, Ohno, N, Yamaguchi, Y. | | Deposit date: | 2010-11-22 | | Release date: | 2011-06-22 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural insights into recognition of triple-helical beta-glucans by an insect fungal receptor
J.Biol.Chem., 286, 2011
|
|
3ACC
 
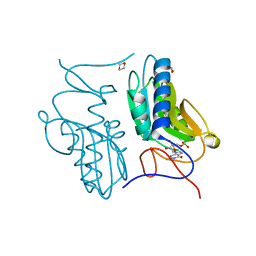 | | Crystal structure of hypoxanthine-guanine phosphoribosyltransferase with GMP from Thermus thermophilus HB8 | | Descriptor: | 1,4-DIETHYLENE DIOXIDE, GUANOSINE-5'-MONOPHOSPHATE, Hypoxanthine-guanine phosphoribosyltransferase | | Authors: | Kanagawa, M, Baba, S, Hirotsu, K, Kuramitsu, S, Yokoyama, S, Kawai, G, Sampei, G, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2009-12-30 | | Release date: | 2010-02-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | Structures of hypoxanthine-guanine phosphoribosyltransferase (TTHA0220) from Thermus thermophilus HB8.
Acta Crystallogr.,Sect.F, 66, 2010
|
|
3ACB
 
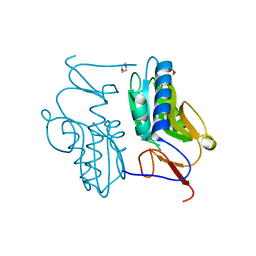 | | Crystal structure of hypoxanthine-guanine phosphoribosyltransferase from Thermus thermophilus HB8 | | Descriptor: | 1,4-DIETHYLENE DIOXIDE, Hypoxanthine-guanine phosphoribosyltransferase | | Authors: | Kanagawa, M, Baba, S, Hirotsu, K, Kuramitsu, S, Yokoyama, S, Kawai, G, Sampei, G, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2009-12-30 | | Release date: | 2010-02-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Structures of hypoxanthine-guanine phosphoribosyltransferase (TTHA0220) from Thermus thermophilus HB8.
Acta Crystallogr.,Sect.F, 66, 2010
|
|
1HT9
 
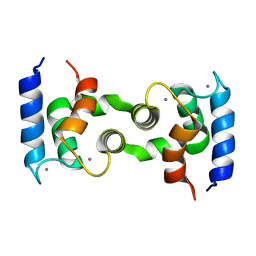 | | DOMAIN SWAPPING EF-HANDS | | Descriptor: | CALBINDIN D9K, CALCIUM ION | | Authors: | Hakansson, M, Svensson, A.L, Fast, J, Linse, S. | | Deposit date: | 2000-12-29 | | Release date: | 2001-05-09 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | An extended hydrophobic core induces EF-hand swapping.
Protein Sci., 10, 2001
|
|
2CV9
 
 | |
4IRM
 
 | |
1EWC
 
 | | CRYSTAL STRUCTURE OF ZN2+ LOADED STAPHYLOCOCCAL ENTEROTOXIN H | | Descriptor: | ENTEROTOXIN H, ZINC ION | | Authors: | Hakansson, M, Petersson, K, Nilsson, H, Forsberg, G, Bjork, P, Antonsson, P, Svensson, A. | | Deposit date: | 2000-04-25 | | Release date: | 2000-05-31 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | The crystal structure of staphylococcal enterotoxin H: implications for binding properties to MHC class II and TcR molecules.
J.Mol.Biol., 302, 2000
|
|
1ENF
 
 | | CRYSTAL STRUCTURE OF STAPHYLOCOCCAL ENTEROTOXIN H DETERMINED TO 1.69 A RESOLUTION | | Descriptor: | ENTEROTOXIN H, SULFATE ION | | Authors: | Hakansson, M, Petersson, K, Nilsson, H, Forsberg, G, Bjork, P, Antonsson, P, Svensson, A. | | Deposit date: | 2000-03-21 | | Release date: | 2000-04-19 | | Last modified: | 2017-10-04 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | The crystal structure of staphylococcal enterotoxin H: implications for binding properties to MHC class II and TcR molecules.
J.Mol.Biol., 302, 2000
|
|
1F77
 
 | | STAPHYLOCOCCAL ENTEROTOXIN H DETERMINED TO 2.4 A RESOLUTION | | Descriptor: | ENTEROTOXIN H, SULFATE ION | | Authors: | Hakansson, M, Petersson, K, Nilsson, H, Forsberg, G, Bjork, P. | | Deposit date: | 2000-06-26 | | Release date: | 2000-07-19 | | Last modified: | 2017-10-04 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The crystal structure of staphylococcal enterotoxin H: implications for binding properties to MHC class II and TcR molecules.
J.Mol.Biol., 302, 2000
|
|
4Z2U
 
 | |
4Z2T
 
 | |
5BRT
 
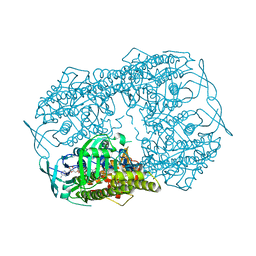 | | Crystal Structure of 2-hydroxybiphenyl 3-monooxygenase from Pseudomonas azelaica with 2-hydroxybiphenyl in the active site | | Descriptor: | 2-HYDROXYBIPHENYL, 2-hydroxybiphenyl-3-monooxygenase, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Kanteev, M, Bregman-Cohen, A, Deri, B, Adir, N, Fishman, A. | | Deposit date: | 2015-06-01 | | Release date: | 2015-08-19 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | A crystal structure of 2-hydroxybiphenyl 3-monooxygenase with bound substrate provides insights into the enzymatic mechanism.
Biochim.Biophys.Acta, 1854, 2015
|
|
5AVM
 
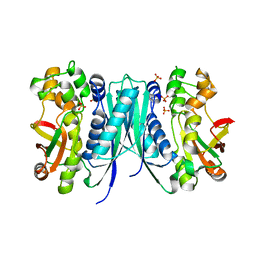 | | Crystal structures of 5-aminoimidazole ribonucleotide (AIR) synthetase, PurM, from Thermus thermophilus | | Descriptor: | Phosphoribosylformylglycinamidine cyclo-ligase, SULFATE ION | | Authors: | Kanagawa, M, Baba, S, Watanabe, Y, Nakagawa, N, Ebihara, A, Sampei, G, Kawai, G, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2015-06-23 | | Release date: | 2015-11-25 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structures and ligand binding of PurM proteins from Thermus thermophilus and Geobacillus kaustophilus
J.Biochem., 159, 2016
|
|
8R74
 
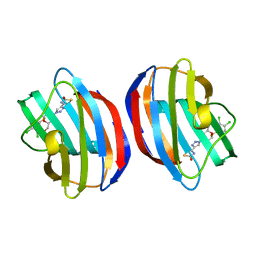 | | Galectin-1 in complex with thiogalactoside derivative | | Descriptor: | (2~{R},3~{R},4~{S},5~{R},6~{R})-2-(3,4-dichlorophenyl)sulfanyl-6-(hydroxymethyl)-4-[4-(2-oxidanyl-1,3-thiazol-4-yl)-1,2,3-triazol-1-yl]oxane-3,5-diol, Galectin-1 | | Authors: | Hakansson, M, Diehl, C, Peterson, K, Zetterberg, F, Nilsson, U. | | Deposit date: | 2023-11-23 | | Release date: | 2024-06-05 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Discovery of the Selective and Orally Available Galectin-1 Inhibitor GB1908 as a Potential Treatment for Lung Cancer.
J.Med.Chem., 67, 2024
|
|
6Y78
 
 | | Structure of galectin-3C in complex with lactose determined by serial crystallography using a silicon nitride membrane support | | Descriptor: | Galectin-3, beta-D-galactopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Hakansson, M, Welin, M, Shilova, A, Kovacic, R, Mueller, U, Logan, D.T. | | Deposit date: | 2020-02-28 | | Release date: | 2020-07-29 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Current status and future opportunities for serial crystallography at MAX IV Laboratory.
J.Synchrotron Radiat., 27, 2020
|
|
7OLY
 
 | | Structure of activin A in complex with an ActRIIB-Alk4 fusion reveal insight into activin receptor interactions | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-6)-alpha-D-mannopyranose-(1-6)-[alpha-D-mannopyranose-(1-3)]alpha-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Activin receptor type-1B, ... | | Authors: | Hakansson, M, Rose, N.C, Castonguay, R, Logan, D.T, Krishnan, L. | | Deposit date: | 2021-05-20 | | Release date: | 2022-02-23 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.265 Å) | | Cite: | Structures of activin ligand traps using natural sets of type I and type II TGF beta receptors.
Iscience, 25, 2022
|
|
6ET4
 
 | | HUMAN DIHYDROOROTATE DEHYDROGENASE IN COMPLEX WITH NOVEL INHIBITOR | | Descriptor: | (4S)-2,6-DIOXOHEXAHYDROPYRIMIDINE-4-CARBOXYLIC ACID, ACETIC ACID, CHLORIDE ION, ... | | Authors: | Hakansson, M, Walse, B, Gustavsson, A.-L, Lain, S. | | Deposit date: | 2017-10-25 | | Release date: | 2018-03-28 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | A DHODH inhibitor increases p53 synthesis and enhances tumor cell killing by p53 degradation blockage.
Nat Commun, 9, 2018
|
|
6SU5
 
 | | Ph2119 endolysin from Thermus scotoductus MAT2119 bacteriophage Ph2119 | | Descriptor: | GLYCEROL, Lysozyme, PHOSPHATE ION, ... | | Authors: | Hakansson, M, Al-Karadaghi, S, Plotka, M, Kaczorowska, A.K, Kaczorowski, T. | | Deposit date: | 2019-09-13 | | Release date: | 2020-09-30 | | Last modified: | 2021-04-14 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Molecular Characterization of a Novel Lytic Enzyme LysC from Clostridium intestinale URNW and Its Antibacterial Activity Mediated by Positively Charged N -Terminal Extension.
Int J Mol Sci, 21, 2020
|
|
6SSC
 
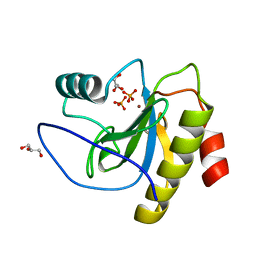 | | N-acetylmuramoyl-L-alanine amidase LysC from Clostridium intestinale URNW | | Descriptor: | GLYCEROL, N-acetylmuramoyl-L-alanine amidase, PHOSPHATE ION, ... | | Authors: | Hakansson, M, Al-Karadaghi, S, Kovacic, R, Plotka, M, Kaczorowska, A.K, Kaczorowski, T. | | Deposit date: | 2019-09-06 | | Release date: | 2020-09-30 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.21 Å) | | Cite: | Molecular Characterization of a Novel Lytic Enzyme LysC from Clostridium intestinale URNW and Its Antibacterial Activity Mediated by Positively Charged N -Terminal Extension.
Int J Mol Sci, 21, 2020
|
|
5I38
 
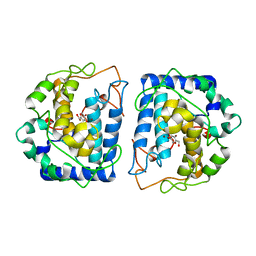 | | Crystal Structure of tyrosinase from Bacillus megaterium with inhibitor kojic acid in the active site | | Descriptor: | 5-HYDROXY-2-(HYDROXYMETHYL)-4H-PYRAN-4-ONE, COPPER (II) ION, Tyrosinase | | Authors: | Kanteev, M, Goldfeder, M, Deri, B, Adir, N, Fishman, A. | | Deposit date: | 2016-02-10 | | Release date: | 2016-10-12 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The unravelling of the complex pattern of tyrosinase inhibition.
Sci Rep, 6, 2016
|
|
6SRT
 
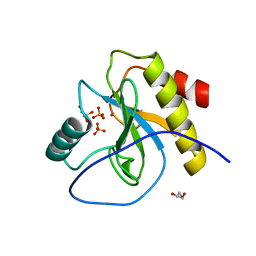 | | Endolysine N-acetylmuramoyl-L-alanine amidase LysCS from Clostridium intestinale URNW | | Descriptor: | GLYCEROL, N-acetylmuramoyl-L-alanine amidase, PHOSPHATE ION, ... | | Authors: | Hakansson, M, Al-Karadaghi, S, Plotka, M, Kaczorowska, A.-K, Kaczorowski, T. | | Deposit date: | 2019-09-06 | | Release date: | 2020-09-30 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.21 Å) | | Cite: | Structure and function of endolysines LysCS, LysC from Clostridium intestinale
To Be Published
|
|
