2LDD
 
 | | Solution structure of the estrogen receptor-binding stapled peptide SP6 (Ac-EKHKILXRLLXDS-NH2) | | Descriptor: | Estrogen receptor-binding stapled peptide SP6 | | Authors: | Phillips, C, Bazin, R, Bent, A, Davies, N, Moore, R, Pannifer, A, Pickford, A, Prior, S, Read, C, Roberts, L, Schade, M, Scott, A, Brown, D, Xu, B, Irving, S. | | Deposit date: | 2011-05-21 | | Release date: | 2011-07-06 | | Last modified: | 2024-11-06 | | Method: | SOLUTION NMR | | Cite: | Design and structure of stapled peptides binding to estrogen receptors.
J.Am.Chem.Soc., 133, 2011
|
|
2LDC
 
 | | Solution structure of the estrogen receptor-binding stapled peptide SP1 (Ac-HXILHXLLQDS-NH2) | | Descriptor: | Estrogen receptor-binding stapled peptide SP1 | | Authors: | Phillips, C, Bazin, R, Bent, A, Davies, N, Moore, R, Pannifer, A, Pickford, A, Prior, S, Read, C, Roberts, L, Schade, M, Scott, A, Brown, D, Xu, B, Irving, S. | | Deposit date: | 2011-05-20 | | Release date: | 2011-07-06 | | Last modified: | 2023-11-15 | | Method: | SOLUTION NMR | | Cite: | Design and structure of stapled peptides binding to estrogen receptors.
J.Am.Chem.Soc., 133, 2011
|
|
2LDA
 
 | | Solution structure of the estrogen receptor-binding stapled peptide SP2 (Ac-HKXLHQXLQDS-NH2) | | Descriptor: | Estrogen receptor-binding stapled peptide SP2 | | Authors: | Phillips, C, Bazin, R, Bent, A, Davies, N, Moore, R, Pannifer, A, Pickford, A, Prior, S, Read, C, Roberts, L, Schade, M, Scott, A, Brown, D, Xu, B, Irving, S. | | Deposit date: | 2011-05-20 | | Release date: | 2011-07-06 | | Last modified: | 2024-11-06 | | Method: | SOLUTION NMR | | Cite: | Design and structure of stapled peptides binding to estrogen receptors.
J.Am.Chem.Soc., 133, 2011
|
|
7T1I
 
 | | Crystal structure of CAB1 Pantothenate Kinase from Saccharomyces cerevisiae in complex with compound YU385597 | | Descriptor: | (8S)-2-{[(4-tert-butylphenyl)methyl]amino}-5-[(piperidin-1-yl)methyl][1,2,4]triazolo[1,5-a]pyrimidin-7(6H)-one, 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Gihaz, S, Ben Mamoun, C. | | Deposit date: | 2021-12-02 | | Release date: | 2022-10-05 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | High-resolution crystal structure and chemical screening reveal pantothenate kinase as a new target for antifungal development.
Structure, 30, 2022
|
|
7T1H
 
 | | Crystal structure of CAB1 Pantothenate Kinase from Saccharomyces cerevisiae in complex with compound YU281445 | | Descriptor: | (8R)-2-{[(4-tert-butylphenyl)methyl]amino}-5-[(morpholin-4-yl)methyl][1,2,4]triazolo[1,5-a]pyrimidin-7(4H)-one, GLYCEROL, Pantothenate kinase CAB1 | | Authors: | Gihaz, S, Ben Mamoun, C. | | Deposit date: | 2021-12-01 | | Release date: | 2022-10-05 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | High-resolution crystal structure and chemical screening reveal pantothenate kinase as a new target for antifungal development.
Structure, 30, 2022
|
|
7T1G
 
 | | Crystal structure of CAB1 Pantothenate Kinase from Saccharomyces cerevisiae in complex with compound YU385595 | | Descriptor: | (8S)-N~2~-[(4-tert-butylphenyl)methyl]-N~7~,N~7~-dimethyl-5-[(morpholin-4-yl)methyl][1,2,4]triazolo[1,5-a]pyrimidine-2,7-diamine, Pantothenate kinase CAB1 | | Authors: | Gihaz, S, Ben Mamoun, C. | | Deposit date: | 2021-12-01 | | Release date: | 2022-10-05 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | High-resolution crystal structure and chemical screening reveal pantothenate kinase as a new target for antifungal development.
Structure, 30, 2022
|
|
4AUA
 
 | | Liganded X-ray crystal structure of cyclin dependent kinase 6 (CDK6) | | Descriptor: | 1H-benzimidazol-2-yl(1H-pyrrol-2-yl)methanone, CYCLIN-DEPENDENT KINASE 6 | | Authors: | Cho, Y.S, Angove, H, Brain, C, Chen, C.H.T, Cheng, R, Chopra, R, Chung, K, Congreve, M, Dagostin, C, Davis, D, Feltell, R, Giraldes, J, Hiscock, S, Kim, S, Kovats, S, Lagu, B, Lewry, K, Loo, A, Lu, Y, Luzzio, M, Maniara, W, Mcmenamin, R, Mortenson, P, Benning, R, O'Reilly, M, Rees, D, Shen, J, Smith, T, Wang, Y, Williams, G, Woolford, A, Wrona, W, Xu, M, Yang, F, Howard, S. | | Deposit date: | 2012-05-15 | | Release date: | 2013-02-06 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Fragment-Based Discovery of 7-Azabenzimidazoles as Potent, Highly Selective, and Orally Active CDK4/6 Inhibitors.
ACS Med Chem Lett, 3, 2012
|
|
7O4O
 
 | | Structure of Staphylococcus aureus m1A22-tRNA methyltransferase in complex with S-adenosylhomocysteine | | Descriptor: | GLYCEROL, S-ADENOSYL-L-HOMOCYSTEINE, tRNA (Adenine(22)-N(1))-methyltransferase | | Authors: | Gloster, T.M, Czekster, C.M, da Silva, R.G. | | Deposit date: | 2021-04-06 | | Release date: | 2022-04-06 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Structure, dynamics, and molecular inhibition of the Staphylococcus aureus m 1 A22-tRNA methyltransferase TrmK.
J.Biol.Chem., 298, 2022
|
|
7O4N
 
 | | Structure of Staphylococcus aureus m1A22-tRNA methyltransferase in complex with S-adenosylmethionine | | Descriptor: | GLYCEROL, S-ADENOSYLMETHIONINE, tRNA (Adenine(22)-N(1))-methyltransferase | | Authors: | Gloster, T.M, Czekster, C.M, da Silva, R.G. | | Deposit date: | 2021-04-06 | | Release date: | 2022-04-20 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structure, dynamics, and molecular inhibition of the Staphylococcus aureus m 1 A22-tRNA methyltransferase TrmK.
J.Biol.Chem., 298, 2022
|
|
7O4M
 
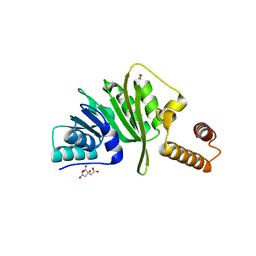 | | Structure of Staphylococcus aureus m1A22-tRNA methyltransferase | | Descriptor: | CITRIC ACID, GLYCEROL, tRNA (Adenine(22)-N(1))-methyltransferase | | Authors: | Gloster, T.M, Czekster, C.M, da Silva, R.G. | | Deposit date: | 2021-04-06 | | Release date: | 2022-04-27 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structure, dynamics, and molecular inhibition of the Staphylococcus aureus m 1 A22-tRNA methyltransferase TrmK.
J.Biol.Chem., 298, 2022
|
|
8VSG
 
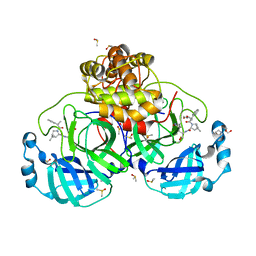 | | SARS-CoV-2 main protease with covalent inhibitor | | Descriptor: | (1R,2S,5S)-N-{(2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-(1-phenylcyclopropane-1-carbonyl)-3-azabicyclo[3.1.0]hexane-2-carboxamide, 1,2-ETHANEDIOL, 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ... | | Authors: | Bell, J.A, Bandera, A.M. | | Deposit date: | 2024-01-24 | | Release date: | 2024-04-03 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.071 Å) | | Cite: | Exploiting high-energy hydration sites for the discovery of potent peptide aldehyde inhibitors of the SARS-CoV-2 main protease with cellular antiviral activity.
Bioorg.Med.Chem., 103, 2024
|
|
8VQX
 
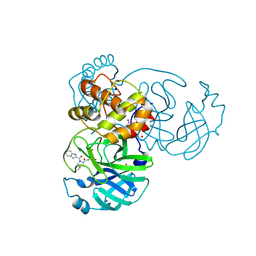 | | Structure of SARS-CoV-2 main protease with potent peptide aldehyde inhibitor | | Descriptor: | 1,2-ETHANEDIOL, 3C-like proteinase nsp5, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Dougan, D.R, Lane, W. | | Deposit date: | 2024-01-19 | | Release date: | 2024-08-07 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Exploiting high-energy hydration sites for the discovery of potent peptide aldehyde inhibitors of the SARS-CoV-2 main protease with cellular antiviral activity.
Bioorg.Med.Chem., 103, 2024
|
|
5I7U
 
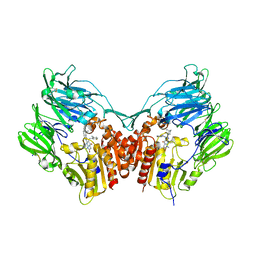 | | Human DPP4 in complex with a novel tricyclic hetero-cycle inhibitor | | Descriptor: | 2-({2-[(3R)-3-aminopiperidin-1-yl]-5-methyl-6,9-dioxo-5,6,7,9-tetrahydro-1H-imidazo[1,2-a]purin-1-yl}methyl)-4-fluorobenzonitrile, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Scapin, G. | | Deposit date: | 2016-02-18 | | Release date: | 2016-06-08 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Discovery of Novel Tricyclic Heterocycles as Potent and Selective DPP-4 Inhibitors for the Treatment of Type 2 Diabetes.
Acs Med.Chem.Lett., 7, 2016
|
|
4NCD
 
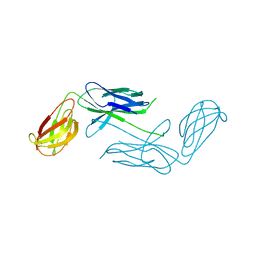 | | Crystal Structure of Class 5 Fimbriae Chaperone CfaA | | Descriptor: | Gram-negative pili assembly chaperone, N-terminal domain protein | | Authors: | Bao, R, Xia, D. | | Deposit date: | 2013-10-24 | | Release date: | 2014-08-27 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.037 Å) | | Cite: | Structure of CfaA Suggests a New Family of Chaperones Essential for Assembly of Class 5 Fimbriae.
Plos Pathog., 10, 2014
|
|
4R91
 
 | | BACE-1 in complex with (R)-4-(2-cyclohexylethyl)-4-(((1S,3R)-3-(cyclopentylamino)cyclohexyl)methyl)-1-methyl-5-oxoimidazolidin-2-iminium | | Descriptor: | (2E,5R)-5-(2-cyclohexylethyl)-5-{[(1S,3R)-3-(cyclopentylamino)cyclohexyl]methyl}-2-imino-3-methylimidazolidin-4-one, Beta-secretase 1, L(+)-TARTARIC ACID | | Authors: | Orth, P, Caldwell, J.P, Strickland, C. | | Deposit date: | 2014-09-03 | | Release date: | 2014-11-05 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Discovery of potent iminoheterocycle BACE1 inhibitors.
Bioorg.Med.Chem.Lett., 24, 2014
|
|
4R95
 
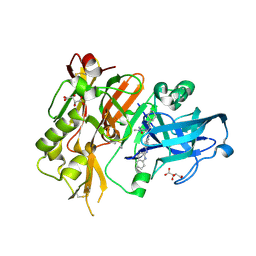 | | BACE-1 in complex with 2-(((1R,3S)-3-(((R)-4-(2-cyclohexylethyl)-2-iminio-1-methyl-5-oxoimidazolidin-4-yl)methyl)cyclohexyl)amino)quinolin-1-ium | | Descriptor: | (2E,5R)-5-(2-cyclohexylethyl)-2-imino-3-methyl-5-{[(1S,3R)-3-(quinolin-2-ylamino)cyclohexyl]methyl}imidazolidin-4-one, Beta-secretase 1, L(+)-TARTARIC ACID | | Authors: | Orth, P, Strickland, C, Caldwell, J.P. | | Deposit date: | 2014-09-03 | | Release date: | 2014-11-05 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Discovery of potent iminoheterocycle BACE1 inhibitors.
Bioorg.Med.Chem.Lett., 24, 2014
|
|
4R92
 
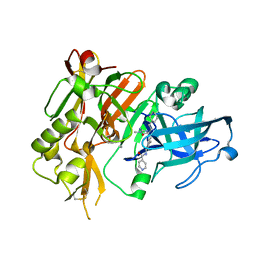 | | BACE-1 in complex with (R)-4-(2-cyclohexylethyl)-4-(((1S,3R)-3-(isonicotinamido)cyclohexyl)methyl)-1-methyl-5-oxoimidazolidin-2-iminium | | Descriptor: | Beta-secretase 1, L(+)-TARTARIC ACID, N-[(1R,3S)-3-{[(2E,4R)-4-(2-cyclohexylethyl)-2-imino-1-methyl-5-oxoimidazolidin-4-yl]methyl}cyclohexyl]pyridine-4-carboxamide | | Authors: | Orth, P, Strickland, C, Caldwell, J.P. | | Deposit date: | 2014-09-03 | | Release date: | 2014-11-05 | | Last modified: | 2014-12-17 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Discovery of potent iminoheterocycle BACE1 inhibitors.
Bioorg.Med.Chem.Lett., 24, 2014
|
|
4R8Y
 
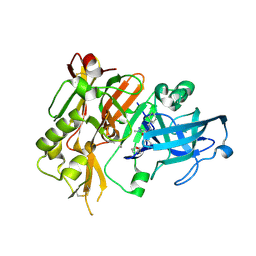 | | BACE-1 in complex with (R)-4-(2-cyclohexylethyl)-4-(((R)-1-(2-cyclopentylacetyl)pyrrolidin-3-yl)methyl)-1-methyl-5-oxoimidazolidin-2-iminium | | Descriptor: | (2E,5R)-5-(2-cyclohexylethyl)-5-{[(3R)-1-(cyclopentylacetyl)pyrrolidin-3-yl]methyl}-2-imino-3-methylimidazolidin-4-one, Beta-secretase 1, L(+)-TARTARIC ACID | | Authors: | Orth, P, Caldwell, J.P, Strickland, C. | | Deposit date: | 2014-09-03 | | Release date: | 2014-11-05 | | Last modified: | 2014-12-17 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Discovery of potent iminoheterocycle BACE1 inhibitors.
Bioorg.Med.Chem.Lett., 24, 2014
|
|
4R93
 
 | | BACE-1 in complex with (R)-4-(2-cyclohexylethyl)-1-methyl-5-oxo-4-(((1S,3R)-3-(3-phenylureido)cyclohexyl)methyl)imidazolidin-2-iminium | | Descriptor: | 1-[(1R,3S)-3-{[(2E,4R)-4-(2-cyclohexylethyl)-2-imino-1-methyl-5-oxoimidazolidin-4-yl]methyl}cyclohexyl]-3-phenylurea, Beta-secretase 1, L(+)-TARTARIC ACID | | Authors: | Orth, P, Strickland, C, Caldwell, J.P. | | Deposit date: | 2014-09-03 | | Release date: | 2014-11-05 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Discovery of potent iminoheterocycle BACE1 inhibitors.
Bioorg.Med.Chem.Lett., 24, 2014
|
|
3L5B
 
 | | Structure of BACE Bound to SCH713601 | | Descriptor: | (2Z,5R)-3-(3-chlorobenzyl)-2-imino-5-methyl-5-(2-methylpropyl)imidazolidin-4-one, Beta-secretase 1, D(-)-TARTARIC ACID | | Authors: | Strickland, C, Zhu, Z. | | Deposit date: | 2009-12-21 | | Release date: | 2010-02-16 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Discovery of Cyclic Acylguanidines as Highly Potent and Selective beta-Site Amyloid Cleaving Enzyme (BACE) Inhibitors: Part I-Inhibitor Design and Validation
J.Med.Chem., 53, 2010
|
|
3L59
 
 | | Structure of BACE Bound to SCH710413 | | Descriptor: | (2Z)-3-(3-chlorobenzyl)-2-imino-5,5-dimethylimidazolidin-4-one, Beta-secretase 1, D(-)-TARTARIC ACID | | Authors: | Strickland, C, Zhu, Z. | | Deposit date: | 2009-12-21 | | Release date: | 2010-02-16 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Discovery of Cyclic Acylguanidines as Highly Potent and Selective beta-Site Amyloid Cleaving Enzyme (BACE) Inhibitors: Part I-Inhibitor Design and Validation
J.Med.Chem., 53, 2010
|
|
3L58
 
 | | Structure of BACE Bound to SCH589432 | | Descriptor: | Beta-secretase 1, N'-{(1S,2R)-1-(3,5-DIFLUOROBENZYL)-2-HYDROXY-3-[(3-METHOXYBENZYL)AMINO]PROPYL}-5-METHYL-N,N-DIPROPYLISOPHTHALAMIDE | | Authors: | Strickland, C, Zhu, Z. | | Deposit date: | 2009-12-21 | | Release date: | 2010-02-16 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Discovery of Cyclic Acylguanidines as Highly Potent and Selective beta-Site Amyloid Cleaving Enzyme (BACE) Inhibitors: Part I-Inhibitor Design and Validation
J.Med.Chem., 53, 2010
|
|
3LPI
 
 | | Structure of BACE Bound to SCH745132 | | Descriptor: | Beta-secretase 1, L(+)-TARTARIC ACID, N'-{(1S,2S)-1-(3,5-difluorobenzyl)-2-hydroxy-2-[(2R)-4-(phenylsulfonyl)piperazin-2-yl]ethyl}-5-methyl-N,N-dipropylbenzene-1,3-dicarboxamide | | Authors: | Strickland, C, Cumming, J. | | Deposit date: | 2010-02-05 | | Release date: | 2010-04-14 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Piperazine sulfonamide BACE1 inhibitors: design, synthesis, and in vivo characterization.
Bioorg.Med.Chem.Lett., 20, 2010
|
|
3L5E
 
 | | Structure of BACE Bound to SCH736062 | | Descriptor: | (4S)-1-(4-{[(2Z,4R)-4-(2-cyclohexylethyl)-4-(cyclohexylmethyl)-2-imino-5-oxoimidazolidin-1-yl]methyl}benzyl)-4-propylimidazolidin-2-one, Beta-secretase 1, D(-)-TARTARIC ACID | | Authors: | Strickland, C, Zhu, Z. | | Deposit date: | 2009-12-21 | | Release date: | 2010-02-16 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Discovery of Cyclic Acylguanidines as Highly Potent and Selective beta-Site Amyloid Cleaving Enzyme (BACE) Inhibitors: Part I-Inhibitor Design and Validation
J.Med.Chem., 53, 2010
|
|
3LPK
 
 | | Structure of BACE Bound to SCH747123 | | Descriptor: | Beta-secretase 1, L(+)-TARTARIC ACID, N-[(1S,2S)-1-(3,5-difluorobenzyl)-2-hydroxy-2-{(2R)-4-[(3-methylphenyl)sulfonyl]piperazin-2-yl}ethyl]-3-{[(2R)-2-(methoxymethyl)pyrrolidin-1-yl]carbonyl}-5-methylbenzamide | | Authors: | Strickland, C, Cumming, J. | | Deposit date: | 2010-02-05 | | Release date: | 2010-04-14 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Piperazine sulfonamide BACE1 inhibitors: design, synthesis, and in vivo characterization.
Bioorg.Med.Chem.Lett., 20, 2010
|
|
