2L7B
 
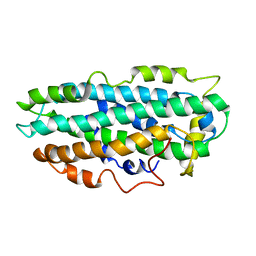 | | NMR Structure of full length apoE3 | | Descriptor: | Apolipoprotein E | | Authors: | Chen, J, Wang, J. | | Deposit date: | 2010-12-07 | | Release date: | 2011-08-03 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Topology of human apolipoprotein E3 uniquely regulates its diverse biological functions.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
2LEM
 
 | |
8J5Y
 
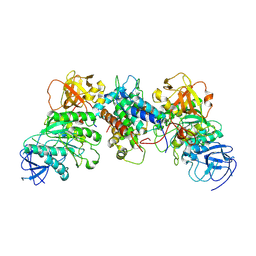 | | Structural and mechanistic insight into ribosomal ITS2 RNA processing by nuclease-kinase machinery | | Descriptor: | LAS1 isoform 1, Polynucleotide 5'-hydroxyl-kinase GRC3 | | Authors: | Chen, J, Chen, H, Li, S, Lin, X, Hu, R, Zhang, K, Liu, L. | | Deposit date: | 2023-04-24 | | Release date: | 2024-01-17 | | Method: | ELECTRON MICROSCOPY (3.07 Å) | | Cite: | Structural and mechanistic insights into ribosomal ITS2 RNA processing by nuclease-kinase machinery.
Elife, 12, 2024
|
|
8J60
 
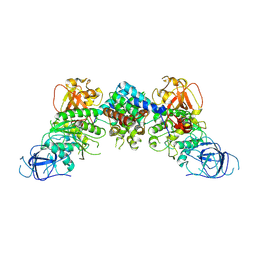 | | Structural and mechanistic insight into ribosomal ITS2 RNA processing by nuclease-kinase machinery | | Descriptor: | LAS1 protein, Polynucleotide 5'-hydroxyl-kinase GRC3 | | Authors: | Chen, J, Chen, H, Li, S, Lin, X, Hu, R, Zhang, K, Liu, L. | | Deposit date: | 2023-04-24 | | Release date: | 2024-01-17 | | Method: | ELECTRON MICROSCOPY (3.39 Å) | | Cite: | Structural and mechanistic insights into ribosomal ITS2 RNA processing by nuclease-kinase machinery.
Elife, 12, 2024
|
|
1EY6
 
 | |
1EYD
 
 | |
1EYC
 
 | |
1EY0
 
 | |
1EY8
 
 | |
1EY7
 
 | |
1EY4
 
 | |
1EYA
 
 | |
1EY5
 
 | |
1EY9
 
 | |
1EZ8
 
 | |
1EZ6
 
 | |
1QU1
 
 | | CRYSTAL STRUCTURE OF EHA2 (23-185) | | Descriptor: | PROTEIN (INFLUENZA RECOMBINANT HA2 CHAIN) | | Authors: | Chen, J, Skehel, J.J, Wiley, D.C. | | Deposit date: | 1999-07-05 | | Release date: | 2000-01-05 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | N- and C-terminal residues combine in the fusion-pH influenza hemagglutinin HA(2) subunit to form an N cap that terminates the triple-stranded coiled coil.
Proc.Natl.Acad.Sci.USA, 96, 1999
|
|
5XM5
 
 | |
5YHQ
 
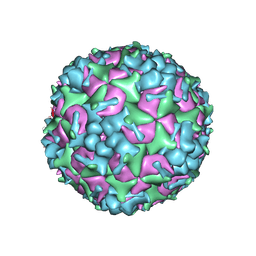 | | Cryo-EM Structure of CVA6 VLP | | Descriptor: | Capsid protein VP1, Capsid protein VP3, capsid protein VP0 | | Authors: | Chen, J, Zhang, C, Huang, Z, Cong, Y. | | Deposit date: | 2017-09-29 | | Release date: | 2017-10-25 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | A 3.0-Angstrom Resolution Cryo-Electron Microscopy Structure and Antigenic Sites of Coxsackievirus A6-Like Particles.
J. Virol., 92, 2018
|
|
5H11
 
 | |
5I65
 
 | |
5VT0
 
 | |
5Y9Q
 
 | |
5YXC
 
 | | Crystal structure of Zinc binding protein ZinT in complex with citrate from E. coli | | Descriptor: | CITRIC ACID, Metal-binding protein ZinT, ZINC ION | | Authors: | Chen, J, Wang, L, Shang, F, Xu, Y. | | Deposit date: | 2017-12-04 | | Release date: | 2017-12-20 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.763 Å) | | Cite: | Crystal structure of E. coli ZinT with one zinc-binding mode and complexed with citrate
Biochem. Biophys. Res. Commun., 500, 2018
|
|
5Z7H
 
 | |
