2HEB
 
 | | CONTRIBUTION OF WATER MOLECULES IN THE INTERIOR OF A PROTEIN TO THE CONFORMATIONAL STABILITY | | Descriptor: | LYSOZYME, SODIUM ION | | Authors: | Takano, K, Funahashi, J, Yamagata, Y, Fujii, S, Yutani, K. | | Deposit date: | 1997-09-16 | | Release date: | 1998-01-28 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Contribution of water molecules in the interior of a protein to the conformational stability.
J.Mol.Biol., 274, 1997
|
|
8Y2S
 
 | | P-hydroxybenzoate hydroxylase complexed with 4-hydroxy-3-methylbenzoic acid | | Descriptor: | 3-methyl-4-oxidanyl-benzoic acid, 4-hydroxybenzoate 3-monooxygenase, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Hara, K, Hashimoto, H, Matsushita, T, Kishimoto, S, Watanabe, K. | | Deposit date: | 2024-01-27 | | Release date: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Functional Enhancement of Flavin-Containing Monooxygenase through Machine Learning Methodology
Acs Catalysis, 14, 2024
|
|
8YWO
 
 | | Crystal structure of L-azetidine-2-carboxylate hydrolase soaked in (S)-azetidine-2-carboxylic acid | | Descriptor: | (2S)-azetidine-2-carboxylic acid, (S)-2-haloacid dehalogenase | | Authors: | Toyoda, M, Mizutani, K, Mikami, B, Wackett, L.P, Esaki, N, Kurihara, T. | | Deposit date: | 2024-03-31 | | Release date: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Research for the crystal structure of L-azetidine-2-carboxylate hydrolase
To Be Published
|
|
8YVW
 
 | | Crystal structure of D12N mutant of L-azetidine-2-carboxylate hydrolase | | Descriptor: | (S)-2-haloacid dehalogenase, FORMIC ACID, IMIDAZOLE, ... | | Authors: | Toyoda, M, Mizutani, K, Mikami, B, Wackett, L.P, Esaki, N, Kurihara, T. | | Deposit date: | 2024-03-29 | | Release date: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.19 Å) | | Cite: | Research for the crystal structure of L-azetidine-2-carboxylate hydrolase
To Be Published
|
|
8X3H
 
 | |
3X3U
 
 | |
2RJ2
 
 | | Crystal Structure of the Sugar Recognizing SCF Ubiquitin Ligase at 1.7 Resolution | | Descriptor: | CHLORIDE ION, F-box only protein 2, NICKEL (II) ION | | Authors: | Vaijayanthimala, S, Velmurugan, D, Mizushima, T, Yamane, T, Yoshida, Y, Tanaka, K. | | Deposit date: | 2007-10-14 | | Release date: | 2008-10-14 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal Structure of the Sugar Recognizing SCF Ubiquitin Ligase at 1.7 Resolution
To be Published
|
|
2HEA
 
 | | CONTRIBUTION OF WATER MOLECULES IN THE INTERIOR OF A PROTEIN TO THE CONFORMATIONAL STABILITY | | Descriptor: | LYSOZYME, SODIUM ION | | Authors: | Takano, K, Funahashi, J, Yamagata, Y, Fujii, S, Yutani, K. | | Deposit date: | 1997-09-16 | | Release date: | 1998-01-14 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Contribution of water molecules in the interior of a protein to the conformational stability.
J.Mol.Biol., 274, 1997
|
|
2HEF
 
 | | CONTRIBUTION OF WATER MOLECULES IN THE INTERIOR OF A PROTEIN TO THE CONFORMATIONAL STABILITY | | Descriptor: | LYSOZYME, SODIUM ION | | Authors: | Takano, K, Funahashi, J, Yamagata, Y, Fujii, S, Yutani, K. | | Deposit date: | 1997-09-16 | | Release date: | 1998-01-14 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Contribution of water molecules in the interior of a protein to the conformational stability.
J.Mol.Biol., 274, 1997
|
|
4IFJ
 
 | | Crystal Structures of apo Keap1, Keap1-peptide, and Keap1-compound complexes | | Descriptor: | Kelch-like ECH-associated protein 1 | | Authors: | Pan, H, Lin, M, Yang, Y, Callaway, K, Baker, J, Diep, L, Yan, J, Tanaka, K, Zhu, Y.L, Konradi, A.W, Jobling, M, Tam, D, Ren, Z, Cheung, H, Bova, M, Riley, B.E, Yao, N, Artis, D.R. | | Deposit date: | 2012-12-14 | | Release date: | 2014-08-20 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structures of apo Keap1, Keap1-peptide, and Keap1-compound complexes
To be Published
|
|
2HEE
 
 | | CONTRIBUTION OF WATER MOLECULES IN THE INTERIOR OF A PROTEIN TO THE CONFORMATIONAL STABILITY | | Descriptor: | LYSOZYME, SODIUM ION | | Authors: | Takano, K, Funahashi, J, Yamagata, Y, Fujii, S, Yutani, K. | | Deposit date: | 1997-09-16 | | Release date: | 1998-01-14 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Contribution of water molecules in the interior of a protein to the conformational stability.
J.Mol.Biol., 274, 1997
|
|
4IFN
 
 | | Crystal Structures of apo Keap1, Keap1-peptide, and Keap1-compound complexes | | Descriptor: | (1R,2R)-2-{[(1S)-1-[(1,3-dioxo-1,3-dihydro-2H-isoindol-2-yl)methyl]-3,4-dihydroisoquinolin-2(1H)-yl]carbonyl}cyclohexanecarboxylic acid, kelch-like ECH-associated protein 1 | | Authors: | Pan, H, Lin, M, Yang, Y, Callaway, K, Baker, J, Diep, L, Yan, J, Tanaka, K, Zhu, Y.L, Konradi, A.W, Jobling, M, Tam, D, Ren, Z, Cheung, H, Bova, M, Riley, B.E, Yao, N, Artis, D.R. | | Deposit date: | 2012-12-14 | | Release date: | 2013-12-18 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structures of apo Keap1, Keap1-peptide, and Keap1-compound complexes
Acta Crystallogr.,Sect.D
|
|
4IFL
 
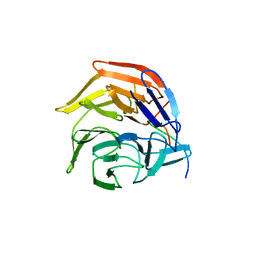 | | Crystal Structures of apo Keap1, Keap1-peptide, and Keap1-compound complexes | | Descriptor: | Nrf2 peptide, kelch-like ECH-associated protein 1 | | Authors: | Pan, H, Lin, M, Yang, Y, Callaway, K, Baker, J, Diep, L, Yan, J, Tanaka, K, Zhu, Y.L, Konradi, A.W, Jobling, M, Tam, D, Ren, Z, Cheung, H, Bova, M, Riley, B.E, Yao, N, Artis, D.R. | | Deposit date: | 2012-12-14 | | Release date: | 2013-12-18 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structures of apo Keap1, Keap1-peptide, and Keap1-compound complexes
Acta Crystallogr.,Sect.D
|
|
1C46
 
 | |
6QIR
 
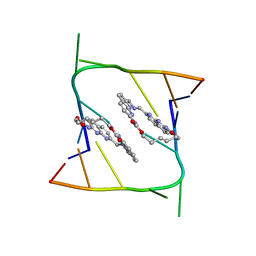 | | Crystal structure of CAG repeats with synthetic CMBL3a compound (model I) | | Descriptor: | CMBL3a, RNA (5'-R(*GP*CP*AP*GP*CP*AP*GP*C)-3') | | Authors: | Kiliszek, A, Blaszczyk, L, Rypniewski, W, Nakatani, K. | | Deposit date: | 2019-01-21 | | Release date: | 2019-09-25 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.531 Å) | | Cite: | Structural insights into synthetic ligands targeting A-A pairs in disease-related CAG RNA repeats.
Nucleic Acids Res., 47, 2019
|
|
5IAU
 
 | |
8WWF
 
 | |
8WWE
 
 | |
8WWD
 
 | |
5INX
 
 | |
5IMH
 
 | |
5IB9
 
 | |
1LHJ
 
 | | ROLE OF PROLINE RESIDUES IN HUMAN LYSOZYME STABILITY: A SCANNING CALORIMETRIC STUDY COMBINED WITH X-RAY STRUCTURE ANALYSIS OF PROLINE MUTANTS | | Descriptor: | HUMAN LYSOZYME | | Authors: | Inaka, K, Matsushima, M, Herning, T, Kuroki, R, Yutani, K, Kikuchi, M. | | Deposit date: | 1992-03-27 | | Release date: | 1994-01-31 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Role of proline residues in human lysozyme stability: a scanning calorimetric study combined with X-ray structure analysis of proline mutants.
Biochemistry, 31, 1992
|
|
1UMI
 
 | | Structural basis of sugar-recognizing ubiquitin ligase | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-alpha-D-glucopyranose, F-box only protein 2 | | Authors: | Mizushima, T, Hirao, T, Yoshida, Y, Lee, S.J, Chiba, T, Iwai, K, Yamaguchi, Y, Kato, K, Tsukihara, T, Tanaka, K. | | Deposit date: | 2003-10-01 | | Release date: | 2004-04-06 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis of sugar-recognizing ubiquitin ligase.
Nat.Struct.Mol.Biol., 11, 2004
|
|
2AEN
 
 | | Crystal structure of the rotavirus strain DS-1 VP8* core | | Descriptor: | ETHANOL, GLYCEROL, Outer capsid protein VP4, ... | | Authors: | Monnier, N, Higo-Moriguchi, K, Sun, Z.-Y.J, Prasad, B.V.V, Taniguchi, K, Dormitzer, P.R. | | Deposit date: | 2005-07-22 | | Release date: | 2006-02-07 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.604 Å) | | Cite: | High-resolution molecular and antigen structure of the VP8*
core of a sialic acid-independent human rotavirus strain
J.Virol., 80, 2006
|
|
