6W9C
 
 | | The crystal structure of papain-like protease of SARS CoV-2 | | Descriptor: | CHLORIDE ION, Non-structural protein 3, ZINC ION | | Authors: | Osipiuk, J, Jedrzejczak, R, Tesar, C, Endres, M, Stols, L, Babnigg, G, Kim, Y, Michalska, K, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-03-22 | | Release date: | 2020-04-01 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The crystal structure of papain-like protease of SARS CoV-2
to be published
|
|
6W5K
 
 | | 1.95 A resolution structure of Norovirus 3CL protease in complex with inhibitor 5g | | Descriptor: | 3C-LIKE PROTEASE, N~2~-{[2-(3-chlorophenyl)-2-methylpropoxy]carbonyl}-N-{(1R,2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]-1-sulfanylpropan-2-yl}-L-leucinamide | | Authors: | Lovell, S, Kashipathy, M.M, Battaile, K.P, Rathnayake, A.D, Kim, Y, Chang, K.O, Groutas, W.C. | | Deposit date: | 2020-03-13 | | Release date: | 2020-09-30 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure-Guided Optimization of Dipeptidyl Inhibitors of Norovirus 3CL Protease.
J.Med.Chem., 63, 2020
|
|
6WBT
 
 | | 2.52 Angstrom Resolution Crystal Structure of 6-phospho-alpha-glucosidase from Gut Microorganisms in Complex with NAD and Glucose-6-phosphate | | Descriptor: | 1,2-ETHANEDIOL, 6-O-phosphono-alpha-D-glucopyranose, MANGANESE (II) ION, ... | | Authors: | Wu, R, Kim, Y, Endres, M, Joachimiak, J. | | Deposit date: | 2020-03-27 | | Release date: | 2021-03-31 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.52 Å) | | Cite: | 2.52 Angstrom Resolution Crystal Structure of 6-phospho-alpha-glucosidase from Gut Microorganisms in Complex with NAD and Glucose-6-phosphate
To Be Published
|
|
6WCF
 
 | | Crystal Structure of ADP ribose phosphatase of NSP3 from SARS-CoV-2 in complex with MES | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Non-structural protein 3 | | Authors: | Michalska, K, Kim, Y, Jedrzejczak, R, Maltseva, N, Endres, M, Mesecar, A, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-03-30 | | Release date: | 2020-04-15 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.065 Å) | | Cite: | Crystal structures of SARS-CoV-2 ADP-ribose phosphatase: from the apo form to ligand complexes.
Iucrj, 7, 2020
|
|
6W5J
 
 | | 1.85 A resolution structure of Norovirus 3CL protease in complex with inhibitor 7d | | Descriptor: | 2-(3-chlorophenyl)-2-methylpropyl [(2S)-3-cyclohexyl-1-({(1R,2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]-1-sulfanylpropan-2-yl}amino)-1-oxopropan-2-yl]carbamate, 3C-LIKE PROTEASE | | Authors: | Lovell, S, Kashipathy, M.M, Battaile, K.P, Rathnayake, A.D, Kim, Y, Chang, K.O, Groutas, W.C. | | Deposit date: | 2020-03-13 | | Release date: | 2020-09-30 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure-Guided Optimization of Dipeptidyl Inhibitors of Norovirus 3CL Protease.
J.Med.Chem., 63, 2020
|
|
6W5L
 
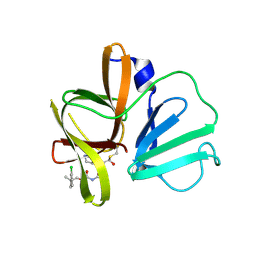 | | 2.1 A resolution structure of Norovirus 3CL protease in complex with inhibitor 7g | | Descriptor: | (2~{S})-~{N}-[(1~{R})-1-[bis($l^{1}-oxidanyl)-methoxy-$l^{5}-sulfanyl]-1-oxidanyl-3-[(3~{R})-2-oxidanylidenepyrrolidin-3-yl]propan-2-yl]-2-[[[2-(3-chlorophenyl)-2-methyl-propoxy]-oxidanylidene-methyl]amino]-4-methyl-pentanamide, 3C-LIKE PROTEASE | | Authors: | Lovell, S, Kashipathy, M.M, Battaile, K.P, Rathnayake, A.D, Kim, Y, Chang, K.O, Groutas, W.C. | | Deposit date: | 2020-03-13 | | Release date: | 2020-09-30 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure-Guided Optimization of Dipeptidyl Inhibitors of Norovirus 3CL Protease.
J.Med.Chem., 63, 2020
|
|
6W02
 
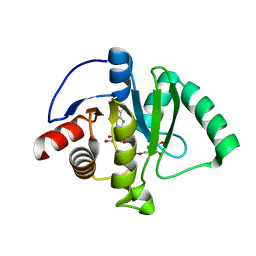 | | Crystal Structure of ADP ribose phosphatase of NSP3 from SARS CoV-2 in the complex with ADP ribose | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE-5-DIPHOSPHORIBOSE, Non-structural protein 3 | | Authors: | Michalska, K, Kim, Y, Jedrzejczak, R, Maltseva, N, Endres, M, Mesecar, A, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-02-28 | | Release date: | 2020-03-11 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structures of SARS-CoV-2 ADP-ribose phosphatase: from the apo form to ligand complexes.
Iucrj, 7, 2020
|
|
6UHE
 
 | |
6UHI
 
 | |
6WTC
 
 | | Crystal Structure of the Second Form of the Co-factor Complex of NSP7 and the C-terminal Domain of NSP8 from SARS CoV-2 | | Descriptor: | ACETIC ACID, Non-structural protein 7, Non-structural protein 8 | | Authors: | Wilamowski, M, Kim, Y, Jedrzejczak, R, Maltseva, N, Endres, M, Godzik, A, Michalska, K, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-05-02 | | Release date: | 2020-05-13 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal Structure of the Second Form of the Co-factor Complex of NSP7 and the C-terminal Domain of NSP8 from SARS CoV-2
To Be Published
|
|
6W4B
 
 | | The crystal structure of Nsp9 RNA binding protein of SARS CoV-2 | | Descriptor: | Non-structural protein 9 | | Authors: | Tan, K, Kim, Y, Jedrzejczak, R, Maltseva, N, Endres, M, Michalska, K, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-03-10 | | Release date: | 2020-03-18 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | The crystal structure of Nsp9 replicase protein of COVID-19
To Be Published
|
|
6W5H
 
 | | 1.85 A resolution structure of Norovirus 3CL protease in complex with inhibitor 5d | | Descriptor: | 2-(3-chlorophenyl)-2-methylpropyl [(2S)-3-cyclohexyl-1-({(2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}amino)-1-oxopropan-2-yl]carbamate, 3C-LIKE PROTEASE | | Authors: | Lovell, S, Kashipathy, M.M, Battaile, K.P, Rathnayake, A.D, Kim, Y, Chang, K.O, Groutas, W.C. | | Deposit date: | 2020-03-13 | | Release date: | 2020-09-30 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure-Guided Optimization of Dipeptidyl Inhibitors of Norovirus 3CL Protease.
J.Med.Chem., 63, 2020
|
|
6WEN
 
 | | Crystal Structure of ADP ribose phosphatase of NSP3 from SARS-CoV-2 in the apo form | | Descriptor: | CHLORIDE ION, Non-structural protein 3 | | Authors: | Michalska, K, Stols, L, Jedrzejczak, R, Endres, M, Babnigg, G, Kim, Y, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-04-02 | | Release date: | 2020-04-15 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Crystal structures of SARS-CoV-2 ADP-ribose phosphatase: from the apo form to ligand complexes.
Iucrj, 7, 2020
|
|
6WRH
 
 | | The crystal structure of Papain-Like Protease of SARS CoV-2 , C111S mutant | | Descriptor: | CHLORIDE ION, GLYCEROL, Non-structural protein 3, ... | | Authors: | Osipiuk, J, Tesar, C, Jedrzejczak, R, Endres, M, Welk, L, Babnigg, G, Kim, Y, Michalska, K, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-04-29 | | Release date: | 2020-05-06 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure of papain-like protease from SARS-CoV-2 and its complexes with non-covalent inhibitors.
Nat Commun, 12, 2021
|
|
5KZV
 
 | | Crystal structure of the xenopus Smoothened cysteine-rich domain (CRD) in complex with 20(S)-hydroxycholesterol | | Descriptor: | (3alpha,8alpha)-cholest-5-ene-3,20-diol, Smoothened | | Authors: | Huang, P, Kim, Y, Salic, A. | | Deposit date: | 2016-07-25 | | Release date: | 2016-08-17 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.616 Å) | | Cite: | Cellular Cholesterol Directly Activates Smoothened in Hedgehog Signaling.
Cell, 166, 2016
|
|
5KZY
 
 | |
5KZZ
 
 | | Crystal structure of the xenopus Smoothened cysteine-rich domain (CRD) in its apo-form | | Descriptor: | ACETATE ION, GLYCEROL, Smoothened, ... | | Authors: | Huang, P, Kim, Y, Salic, A. | | Deposit date: | 2016-07-25 | | Release date: | 2016-08-17 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.332 Å) | | Cite: | Cellular Cholesterol Directly Activates Smoothened in Hedgehog Signaling.
Cell, 166, 2016
|
|
2ARH
 
 | | Crystal Structure of a Protein of Unknown Function AQ1966 from Aquifex aeolicus VF5 | | Descriptor: | CALCIUM ION, SELENIUM ATOM, SULFATE ION, ... | | Authors: | Qiu, Y, Kim, Y, Yang, X, Collart, F, Joachimiak, A, Kossiakoff, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2005-08-19 | | Release date: | 2005-10-04 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (2.46 Å) | | Cite: | Crystal Structure of a Hypothetical Protein Aq_1966 from Aquifex aeolicus VF5
To be Published
|
|
2LNC
 
 | | Solution NMR structure of Norwalk virus protease | | Descriptor: | 3C-like protease | | Authors: | Takahashi, D, Hiromasa, Y, Kim, Y, Anbanandam, A, Chang, K, Prakash, O. | | Deposit date: | 2011-12-22 | | Release date: | 2012-12-26 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structural and dynamics characterization of norovirus protease.
Protein Sci., 22, 2013
|
|
3IGJ
 
 | | Crystal Structure of Maltose O-acetyltransferase Complexed with Acetyl Coenzyme A from Bacillus anthracis | | Descriptor: | ACETYL COENZYME *A, FORMIC ACID, GLYCEROL, ... | | Authors: | Maltseva, N, Kim, Y, Papazisi, L, Anderson, W, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2009-07-27 | | Release date: | 2009-08-04 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal Structure of Maltose O-acetyltransferase Complexed with Acetyl Coenzyme A from Bacillus anthracis
To be Published
|
|
1U14
 
 | | The crystal structure of hypothetical UPF0244 protein yjjX at resolution 1.68 Angstrom | | Descriptor: | Hypothetical UPF0244 protein yjjX, PHOSPHATE ION | | Authors: | Qiu, Y, Kim, Y, Cuff, M, Collart, F, Joachimiak, A, Kossiakoff, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2004-07-14 | | Release date: | 2004-09-21 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | The crystal structure of hypothetical UPF0244 protein yjjX at resolution 1.68 Angstrom
To be Published
|
|
3IQT
 
 | | Structure of the HPT domain of Sensor protein barA from Escherichia coli CFT073. | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CALCIUM ION, Signal transduction histidine-protein kinase barA | | Authors: | Cuff, M.E, Rakowski, E, Kim, Y, Freeman, L, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-08-20 | | Release date: | 2009-09-22 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structure of the HPT domain of Sensor protein barA from Escherichia coli CFT073.
TO BE PUBLISHED
|
|
3HYL
 
 | | Crystal Structure of Transketolase from Bacillus anthracis | | Descriptor: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, FORMIC ACID, ... | | Authors: | Maltseva, N, Kim, Y, Kwon, K, Joachimiak, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2009-06-22 | | Release date: | 2009-06-30 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | Crystal Structure of Transketolase from Bacillus anthracis
To be Published
|
|
1R4V
 
 | | 1.9A crystal structure of protein AQ328 from Aquifex aeolicus | | Descriptor: | CACODYLATE ION, Hypothetical protein AQ_328, ZINC ION | | Authors: | Qiu, Y, Tereshko, V, Kim, Y, Zhang, R, Collart, F, Joachimiak, A, Kossiakoff, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2003-10-08 | | Release date: | 2004-03-30 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The crystal structure of Aq_328 from the hyperthermophilic bacteria Aquifex aeolicus shows an ancestral histone fold.
Proteins, 62, 2006
|
|
3IYD
 
 | | Three-dimensional EM structure of an intact activator-dependent transcription initiation complex | | Descriptor: | ADENOSINE-3',5'-CYCLIC-MONOPHOSPHATE, Catabolite gene activator, DNA (98-MER), ... | | Authors: | Hudson, B.P, Quispe, J, Lara, S, Kim, Y, Berman, H, Arnold, E, Ebright, R.H, Lawson, C.L. | | Deposit date: | 2009-08-01 | | Release date: | 2009-11-10 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (19.799999 Å) | | Cite: | Three-dimensional EM structure of an intact activator-dependent transcription initiation complex
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
