3PTL
 
 | | Crystal structure of proteinase K inhibited by a lactoferrin nonapeptide, Lys-Gly-Glu-Ala-Asp-Ala-Leu-Ser-Leu-Asp at 1.3 A resolution. | | Descriptor: | 10-mer peptide from Lactoferrin, Proteinase K | | Authors: | Shukla, P.K, Sinha, M, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2010-12-03 | | Release date: | 2010-12-22 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Crystal structure of proteinase K inhibited by a lactoferrin nonapeptide, Lys-Gly-Glu-Ala-Asp-Ala-Leu-Ser-Leu-Asp at 1.3 A resolution.
TO BE PUBLISHED
|
|
3QJI
 
 | | Crystal structure of the complex of ribosome inactivating protein with 7-methylguanosine triphosphate at 1.75A resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 7-METHYL-GUANOSINE-5'-TRIPHOSPHATE, Ribosome inactivating protein | | Authors: | Kumar, M, Kushwaha, G.S, Sinha, M, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2011-01-29 | | Release date: | 2011-03-09 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal structure of the complex of ribosome inactivating protein with 7-methylguanosine triphosphate at 1.75A resolution
TO BE PUBLISHED
|
|
3PUE
 
 | | Crystal structure of the complex of Dhydrodipicolinate synthase from Acinetobacter baumannii with lysine at 2.6A resolution | | Descriptor: | Dihydrodipicolinate synthase, GLYCEROL, LYSINE, ... | | Authors: | Jithesh, O, Yamini, S, Kaur, N, Gautam, A, Tewari, R, Kushwaha, G.S, Kaur, P, Srinivasan, A, Sharma, S, Singh, T.P. | | Deposit date: | 2010-12-04 | | Release date: | 2010-12-22 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of the complex of Dhydrodipicolinate synthase from Acinetobacter baumannii with lysine at 2.6A resolution
To be Published
|
|
3Q4Y
 
 | | Crystal structure of group I phospholipase A2 at 2.3 A resolution in 40% ethanol revealed the critical elements of hydrophobicity of the substrate-binding site | | Descriptor: | CALCIUM ION, ETHANOL, Phospholipase A2 isoform 3 | | Authors: | Shukla, P.K, Kaushik, S, Sinha, M, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2010-12-26 | | Release date: | 2011-01-26 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of group I phospholipase A2 at 2.3 A resolution in 40% ethanol revealed the critical elements of hydrophobicity of the substrate-binding site
To be Published
|
|
3Q4P
 
 | | Crystal structure of the complex of type I ribosome inactivating protein with 7n-methyl -8-hydroguanosine-5-p-diphosphate at 1.8 A resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 7N-METHYL-8-HYDROGUANOSINE-5'-DIPHOSPHATE, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Kushwaha, G.S, Yamini, S, Sinha, M, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2010-12-24 | | Release date: | 2011-01-12 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of the complex of type I ribosome inactivating protein with 7n-methyl-8-hydroguanosine-5-p-diphosphate at 1.8 A resolution
To be Published
|
|
3PTN
 
 | | ON THE DISORDERED ACTIVATION DOMAIN IN TRYPSINOGEN. CHEMICAL LABELLING AND LOW-TEMPERATURE CRYSTALLOGRAPHY | | Descriptor: | CALCIUM ION, TRYPSIN | | Authors: | Walter, J, Steigemann, W, Singh, T.P, Bartunik, H, Bode, W, Huber, R. | | Deposit date: | 1981-10-26 | | Release date: | 1982-03-04 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | On the Disordered Activation Domain in Trypsinogen. Chemical Labelling and Low-Temperature Crystallography
Acta Crystallogr.,Sect.B, 38, 1982
|
|
3QS0
 
 | | Crystal structure of the complex of peptidoglycan recognition protein (PGRP-S) with a bound N-acetylglucosamine in the diffusion channel at 2.5 A resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, L(+)-TARTARIC ACID, ... | | Authors: | Dube, D, Sharma, P, Sinha, M, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2011-02-19 | | Release date: | 2011-03-16 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of the complex of peptidoglycan recognition protein (PGRP-S) with a bound N-acetylglucosamine in the diffusion channel AT 2.5 A resolution
To be Published
|
|
3QJ1
 
 | | Crystal structure of camel peptidoglycan recognition protein, PGRP-S with a trapped diethylene glycol in the ligand diffusion channel at 3.2 A resolution | | Descriptor: | DI(HYDROXYETHYL)ETHER, GLYCEROL, L(+)-TARTARIC ACID, ... | | Authors: | Sharma, P, Yamini, S, Sinha, M, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2011-01-28 | | Release date: | 2011-02-16 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Crystal structure of camel peptidoglycan recognition protein, PGRP-S with a trapped diethylene glycol in the ligand diffusion channel at 3.2 A resolution
To be Published
|
|
3RGY
 
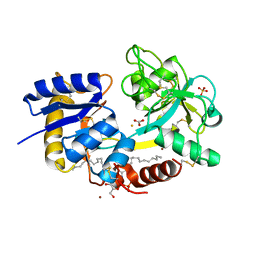 | | Crystal Structure of C-lobe of Bovine lactoferrin Complexed with Lipopolysaccharide at 2.0 A Resolution | | Descriptor: | (R)-((2R,3S,4R,5R,6R)-3-HYDROXY-2-(HYDROXYMETHYL)-5-((R)-3-HYDROXYTETRADECANAMIDO)-6-(PHOSPHONOOXY)TETRAHYDRO-2H-PYRAN-4-YL) 3-HYDROXYTETRADECANOATE, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Shukla, P.K, Sinha, M, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2011-04-11 | | Release date: | 2011-05-04 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of C-lobe of Bovine lactoferrin Complexed with Lipopolysaccharide at 2.0 A Resolution
To be Published
|
|
3PUD
 
 | | Crystal structure of Dhydrodipicolinate synthase from Acinetobacter baumannii at 2.8A resolution | | Descriptor: | Dihydrodipicolinate synthase, GLYCEROL, SULFATE ION | | Authors: | Jithesh, O, Yamini, S, Kaur, N, Gautam, A, Tewari, R, Kushwaha, G.S, Kaur, P, Srinivasan, A, Sharma, S, Singh, T.P. | | Deposit date: | 2010-12-04 | | Release date: | 2010-12-22 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of Dhydrodipicolinate synthase from Acinetobacter baumannii at 2.8A resolution
To be Published
|
|
3SDF
 
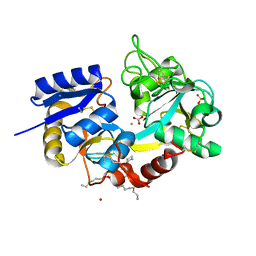 | | Crystal Structure of C-lobe of Bovine lactoferrin Complexed with Lipoteichoic acid at 2.1 A Resolution | | Descriptor: | (2S)-1-({3-O-[2-(acetylamino)-4-amino-2,4,6-trideoxy-beta-D-galactopyranosyl]-alpha-D-glucopyranosyl}oxy)-3-(heptanoyloxy)propan-2-yl (7Z)-pentadec-7-enoate, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Shukla, P.K, Gautam, L, Sinha, M, Bhushan, A, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2011-06-09 | | Release date: | 2011-06-29 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structure of C-lobe of Bovine lactoferrin Complexed with Lipoteichoic acid at 2.1 A Resolution
To be Published
|
|
3OGX
 
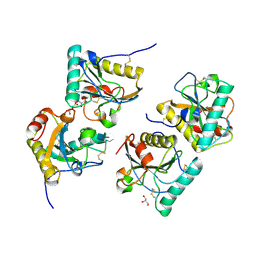 | | Crystal structure of the complex of Peptidoglycan Recognition protein (PGRP-s) with Heparin-Dissacharide at 2.8 A resolution | | Descriptor: | 4-deoxy-2-O-sulfo-alpha-L-threo-hex-4-enopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose, GLYCEROL, L(+)-TARTARIC ACID, ... | | Authors: | Sharma, P, Dube, D, Sinha, M, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2010-08-17 | | Release date: | 2010-09-01 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural basis of heparin binding to camel peptidoglycan recognition protein-S
Int J Biochem Mol Biol, 3, 2012
|
|
3S8H
 
 | | Structure of dihydrodipicolinate synthase complexed with 3-Hydroxypropanoic acid(HPA)at 2.70 A resolution | | Descriptor: | 3-HYDROXY-PROPANOIC ACID, Dihydrodipicolinate synthase | | Authors: | Kumar, M, Kaur, N, Kumar, S, Sinha, M, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2011-05-28 | | Release date: | 2011-06-15 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure of dihydrodipicolinate synthase complexed with 3-Hydroxypropanoic acid(HPA)at 2.70 A resolution
TO BE PUBLISHED
|
|
3OSH
 
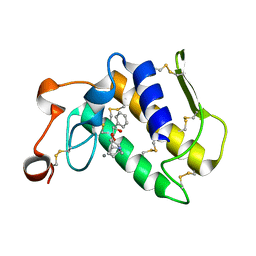 | | Crystal Structure of The Complex of Group 1 Phospholipase A2 With Atropin At 1.5 A Resolution | | Descriptor: | (1R,5S)-8-METHYL-8-AZABICYCLO[3.2.1]OCT-3-YL (2R)-3-HYDROXY-2-PHENYLPROPANOATE, CALCIUM ION, Phospholipase A2 isoform 3 | | Authors: | Shukla, P.K, Kaushik, S, Sinha, M, Bhushan, A, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2010-09-09 | | Release date: | 2010-11-17 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal Structure of The Complex of Group 1 Phospholipase A2 With Atropin At 1.5 A Resolution
To be Published
|
|
3OIH
 
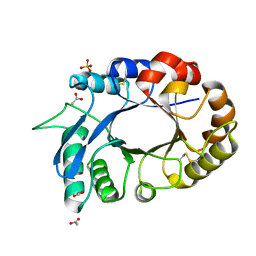 | | Crystal Structure of the complex of xylanase-alpha-amylase inhibitor Protein (XAIP-I) with trehalose at 1.87 A resolution | | Descriptor: | ACETATE ION, Haementhin, PHOSPHATE ION, ... | | Authors: | Kumar, M, Kumar, S, Sinha, M, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2010-08-19 | | Release date: | 2010-09-22 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Crystal Structure of the complex of xylanase-alpha-amylase inhibitor Protein (XAIP-I) with trehalose at 1.87 A resolution
To be Published
|
|
3QV4
 
 | | Crystal structure of the complex of peptidoglycan recognition protein (PGRP-S) with dipeptide L-ALA D-GLU at 2.7 A resolution | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ALANINE, ... | | Authors: | Shukla, P.K, Sharma, P, Sinha, M, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2011-02-25 | | Release date: | 2011-03-30 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of the complex of peptidoglycan recognition protein (PGRP-S) with dipeptide L-ALA D-GLU at 2.7 A resolution
To be Published
|
|
5YHM
 
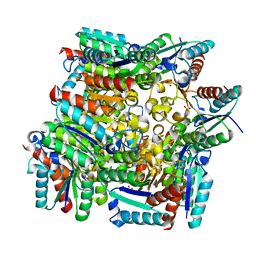 | | Crystal structure of dehydroquinate dehydratase with tris induced oligomerisation at 1.907 Angstrom resolution | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 3-dehydroquinate dehydratase, SULFATE ION | | Authors: | Iqbal, N, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2017-09-28 | | Release date: | 2017-10-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Crystal structure of dehydroquinate dehydratase with tris induced oligomerisation at 1.907 Angstrom resolution
To Be Published
|
|
5YDB
 
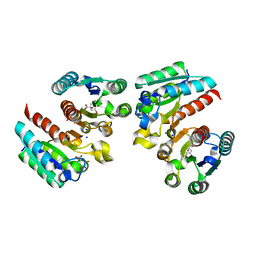 | | Crystal structure of the complex of type II dehydroquinate dehydratase from Acinetobacter baumannii with dehydroquinic acid at 1.76 Angstrom resolution | | Descriptor: | 1,3,4-TRIHYDROXY-5-OXO-CYCLOHEXANECARBOXYLIC ACID, 3-dehydroquinate dehydratase, SODIUM ION | | Authors: | Iqbal, N, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2017-09-12 | | Release date: | 2017-09-20 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Crystal structure of the complex of type II dehydroquinate dehydratase from Acinetobacter baumannii with dehydroquinic acid at 1.76 Angstrom resolution
To Be Published
|
|
5YRR
 
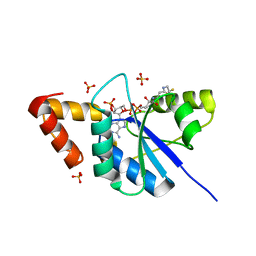 | | The crystal structure of Phosphopantetheine adenylyltransferase from Acinetobacter baumannii with Coenzyme A at 2.88 A resolution | | Descriptor: | COENZYME A, Phosphopantetheine adenylyltransferase, SULFATE ION | | Authors: | Bairagya, H.R, Gupta, A, Iqbal, N, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2017-11-09 | | Release date: | 2017-11-22 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.88 Å) | | Cite: | The crystal structure of Phosphopantetheine adenylyltransferase from Acinetobacter baumannii with Coenzyme A at 2.88 A resolution
To Be Published
|
|
5Z9A
 
 | |
5ZTJ
 
 | | Crystal Structure of GyraseA C-Terminal Domain from Salmonella typhi at 2.4A Resolution | | Descriptor: | DNA gyrase subunit A | | Authors: | Sachdeva, E, Gupta, D, Tiwari, P, Kaur, G, Sharma, S, Singh, T.P, Ethayathulla, A.S, Kaur, P. | | Deposit date: | 2018-05-03 | | Release date: | 2019-05-15 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The pivot point arginines identified in the beta-pinwheel structure of C-terminal domain from Salmonella Typhi DNA Gyrase A subunit.
Sci Rep, 10, 2020
|
|
5ZZV
 
 | | Crystal structure of PEG-1500 crystallized Peptidyl-tRNA Hydrolase from Acinetobacter baumannii at 1.5 A resolution | | Descriptor: | 1,2-ETHANEDIOL, Peptidyl-tRNA hydrolase | | Authors: | Bairagya, H.R, Sharma, P, Iqbal, N, Sharma, S, Singh, T.P. | | Deposit date: | 2018-06-04 | | Release date: | 2018-06-20 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Crystal structure of PEG-1500 crystallized Peptidyl-tRNA Hydrolase from Acinetobacter baumannii at 1.5 A resolution
To Be Published
|
|
6AIE
 
 | | Crystal structure of a new form of RsmD-like RNA methyl transferase from Mycobacterium tuberculosis determined at 1.74 A resolution | | Descriptor: | Putative methyltransferase | | Authors: | Venkataraman, S, Dhankar, A, Sinha, K.M, Manivasakan, P, Iqbal, N, Singh, T.P, Prasad, B.V.L.S. | | Deposit date: | 2018-08-22 | | Release date: | 2018-09-26 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Crystal structure of a new form of RsmD-like RNA methyl transferase from Mycobacterium tuberculosis determined at 1.74 A resolution
To Be Published
|
|
5WV1
 
 | | Crystal structure of the complex of Ribosome inactivating protein from Momordica balsamina with ribose sugar at 1.90 A resolution. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, Ribosome inactivating protein, ... | | Authors: | Shokeen, A, Singh, P.K, Pandey, S, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2016-12-21 | | Release date: | 2017-01-25 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of the complex of Ribosome inactivating protein from Momordica balsamina with ribose sugar at
1.90 A resolution.
To Be Published
|
|
7Y3U
 
 | | Crystal structure of the complex of Lactoperoxidase with Nitric oxide at 2.50A resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, CALCIUM ION, ... | | Authors: | Singh, P.K, Viswanathan, V, Ahmad, N, Rani, C, Sharma, P, Sharma, S, Singh, T.P. | | Deposit date: | 2022-06-13 | | Release date: | 2022-06-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of the complex of Lactoperoxidase with Nitric oxide at 2.50A resolution
To Be Published
|
|
