9K2K
 
 | | Structure of ClpP from Staphylococcus aureus in complex with ZY7 | | Descriptor: | (6~{S},9~{a}~{S})-6-(2-methylpropyl)-8-(naphthalen-1-ylmethyl)-4,7-bis(oxidanylidene)-~{N}-(phenylmethyl)-3,6,9,9~{a}-tetrahydro-2~{H}-pyrazino[1,2-a]pyrimidine-1-carboxamide, ATP-dependent Clp protease proteolytic subunit | | Authors: | Li, J.H, Wu, W, Zhang, T, Yang, C.-G. | | Deposit date: | 2024-10-17 | | Release date: | 2025-02-19 | | Method: | X-RAY DIFFRACTION (2.74 Å) | | Cite: | Structure-Guided Development of ClpP Agonists with Potent Therapeutic Activities against Staphylococcus aureus Infection.
J.Med.Chem., 68, 2025
|
|
7BW6
 
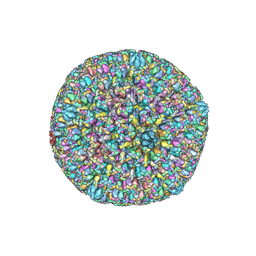 | | Varicella-zoster virus capsid | | Descriptor: | Major capsid protein, Small capsomere-interacting protein, Triplex capsid protein 1, ... | | Authors: | Wang, P.Y, Qi, J.X, Liu, C.C, Sun, J.Q. | | Deposit date: | 2020-04-13 | | Release date: | 2020-09-23 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Cryo-EM structure of the varicella-zoster virus A-capsid.
Nat Commun, 11, 2020
|
|
8HBN
 
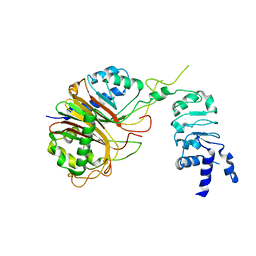 | | Structure of the Mex67-Mtr2-1 heterodimer | | Descriptor: | mRNA export factor MEX67, mRNA transport regulator MTR2 | | Authors: | Li, Z.Q, Chen, S.J, Sui, S.F. | | Deposit date: | 2022-10-29 | | Release date: | 2023-04-12 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.81 Å) | | Cite: | Nuclear export of pre-60S particles through the nuclear pore complex.
Nature, 618, 2023
|
|
8HGK
 
 | | Crystal structure of human ClpP in complex with ZK53 | | Descriptor: | 4-[[3,5-bis(fluoranyl)phenyl]methyl]-N-[(4-bromophenyl)methyl]piperazine-1-carboxamide, ATP-dependent Clp protease proteolytic subunit, mitochondrial, ... | | Authors: | Yang, C.-G, Gan, J.H, Zhou, L.-L. | | Deposit date: | 2022-11-14 | | Release date: | 2023-09-27 | | Last modified: | 2024-04-17 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Selective activator of human ClpP triggers cell cycle arrest to inhibit lung squamous cell carcinoma.
Nat Commun, 14, 2023
|
|
8HFR
 
 | | NPC-trapped pre-60S particle | | Descriptor: | 25S ribosomal RNA, 5.8S ribosomal RNA, 5S ribosomal RNA, ... | | Authors: | Li, Z.Q, Chen, S.J, Sui, S.F. | | Deposit date: | 2022-11-12 | | Release date: | 2023-04-12 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (2.64 Å) | | Cite: | Nuclear export of pre-60S particles through the nuclear pore complex.
Nature, 618, 2023
|
|
8HIF
 
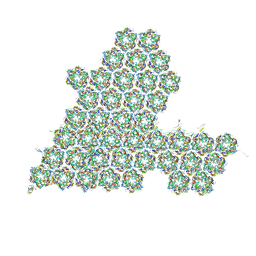 | | One asymmetric unit of Singapore grouper iridovirus capsid | | Descriptor: | Major capsid protein, Penton protein (VP14), VP137, ... | | Authors: | Zhao, Z.N, Liu, C.C, Zhu, D.J, Qi, J.X, Zhang, X.Z, Gao, G.F. | | Deposit date: | 2022-11-20 | | Release date: | 2023-04-19 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Near-atomic architecture of Singapore grouper iridovirus and implications for giant virus assembly.
Nat Commun, 14, 2023
|
|
5Y4K
 
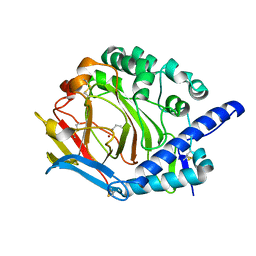 | | Crystal structure of DddY mutant Y260A | | Descriptor: | ACRYLIC ACID, DMSP lyase DddY, ZINC ION | | Authors: | Zhang, Y.Z, Li, C.Y. | | Deposit date: | 2017-08-03 | | Release date: | 2017-11-01 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Mechanistic Insights into Dimethylsulfoniopropionate Lyase DddY, a New Member of the Cupin Superfamily.
J. Mol. Biol., 429, 2017
|
|
5ZK1
 
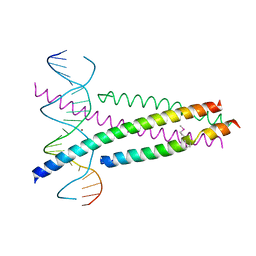 | | Crystal Structure of the CRTC2(SeMet)-CREB-CRE complex | | Descriptor: | CREB-regulated transcription coactivator 2, Cyclic AMP-responsive element-binding protein 1, DNA (5'-D(*CP*TP*TP*GP*GP*CP*TP*GP*AP*CP*GP*TP*CP*AP*GP*CP*CP*AP*AP*G)-3'), ... | | Authors: | Xiang, S, Zhai, L, Valencia-Swain, J. | | Deposit date: | 2018-03-22 | | Release date: | 2018-06-20 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | Structural Insights into the CRTC2-CREB Complex Assembly on CRE.
J. Mol. Biol., 430, 2018
|
|
5ZKO
 
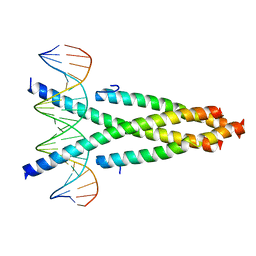 | | Crystal structure of the CRTC2-CREB-CRE complex | | Descriptor: | CREB-regulated transcription coactivator 2, Cyclic AMP-responsive element-binding protein 1, DNA (5'-D(*CP*TP*TP*GP*GP*CP*TP*GP*AP*CP*GP*TP*CP*AP*GP*CP*CP*AP*AP*G)-3') | | Authors: | Xiang, S, Zhai, L, Valecia-Swain, J. | | Deposit date: | 2018-03-24 | | Release date: | 2018-06-20 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | Structural Insights into the CRTC2-CREB Complex Assembly on CRE.
J. Mol. Biol., 430, 2018
|
|
6BMV
 
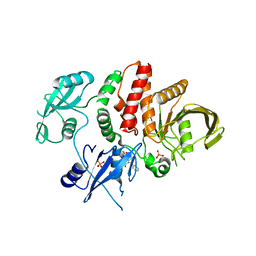 | | Non-receptor Protein Tyrosine Phosphatase SHP2 in Complex with Allosteric Inhibitor SHP504 | | Descriptor: | 3-{4-[(2-chlorophenyl)methyl]-5-oxo-4,5-dihydro[1,2,4]triazolo[4,3-a]quinazolin-1-yl}-4-hydroxybenzoic acid, PHOSPHATE ION, Tyrosine-protein phosphatase non-receptor type 11 | | Authors: | Stams, T, Fodor, M. | | Deposit date: | 2017-11-15 | | Release date: | 2018-01-17 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.053 Å) | | Cite: | Dual Allosteric Inhibition of SHP2 Phosphatase.
ACS Chem. Biol., 13, 2018
|
|
7FJM
 
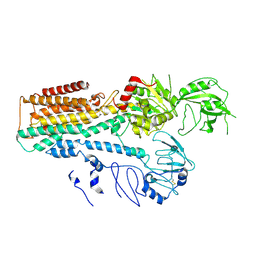 | | Cryo EM structure of lysosomal ATPase | | Descriptor: | Polyamine-transporting ATPase 13A2 | | Authors: | Zhang, S.S. | | Deposit date: | 2021-08-04 | | Release date: | 2023-03-08 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Cryo-EM structures and transport mechanism of human P5B type ATPase ATP13A2.
Cell Discov, 7, 2021
|
|
6KUU
 
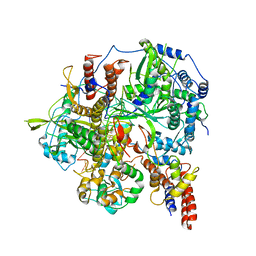 | | Structure of influenza D virus polymerase bound to vRNA promoter in Mode B conformation (Class B3) | | Descriptor: | 3'-vRNA, 5'-vRNA, Polymerase 3, ... | | Authors: | Peng, Q, Peng, R, Qi, J, Gao, G.F, Shi, Y. | | Deposit date: | 2019-09-02 | | Release date: | 2019-12-11 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structure of influenza D virus polymerase bound to vRNA promoter in Mode B conformation (Class B3)
To Be Published
|
|
6BMU
 
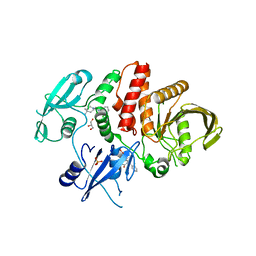 | | Non-receptor Protein Tyrosine Phosphatase SHP2 in Complex with Allosteric Inhibitors SHP099 and SHP244 | | Descriptor: | 4-[(2-chlorophenyl)methyl]-1-(2-hydroxy-3-methoxyphenyl)[1,2,4]triazolo[4,3-a]quinazolin-5(4H)-one, 6-(4-azanyl-4-methyl-piperidin-1-yl)-3-[2,3-bis(chloranyl)phenyl]pyrazin-2-amine, GLYCEROL, ... | | Authors: | Stams, T, Fodor, M. | | Deposit date: | 2017-11-15 | | Release date: | 2018-01-17 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Dual Allosteric Inhibition of SHP2 Phosphatase.
ACS Chem. Biol., 13, 2018
|
|
1J4L
 
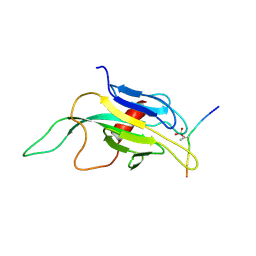 | |
6J5I
 
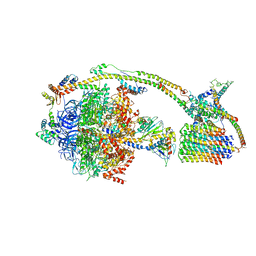 | | Cryo-EM structure of the mammalian DP-state ATP synthase | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP synthase F1 subunit epsilon, ... | | Authors: | Gu, J, Zhang, L, Yi, J, Yang, M. | | Deposit date: | 2019-01-11 | | Release date: | 2019-06-26 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.34 Å) | | Cite: | Cryo-EM structure of the mammalian ATP synthase tetramer bound with inhibitory protein IF1.
Science, 364, 2019
|
|
6J54
 
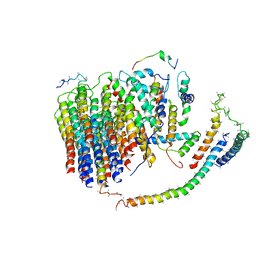 | | Cryo-EM structure of the mammalian E-state ATP synthase FO section | | Descriptor: | ATP synthase membrane subunit 6.8PL, ATP synthase membrane subunit DAPIT, ATP synthase peripheral stalk-membrane subunit b, ... | | Authors: | Gu, J, Zhang, L, Yi, J, Yang, M. | | Deposit date: | 2019-01-10 | | Release date: | 2019-06-26 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.94 Å) | | Cite: | Cryo-EM structure of the mammalian ATP synthase tetramer bound with inhibitory protein IF1.
Science, 364, 2019
|
|
6J5A
 
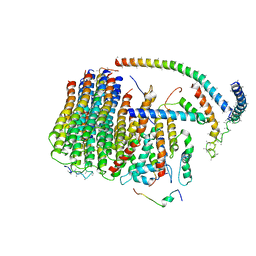 | | Cryo-EM structure of the mammalian DP-state ATP synthase FO section | | Descriptor: | ATP synthase membrane subunit 6.8PL, ATP synthase membrane subunit DAPIT, ATP synthase peripheral stalk-membrane subunit b, ... | | Authors: | Gu, J, Zhang, L, Yi, J, Yang, M. | | Deposit date: | 2019-01-10 | | Release date: | 2019-06-26 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (4.35 Å) | | Cite: | Cryo-EM structure of the mammalian ATP synthase tetramer bound with inhibitory protein IF1.
Science, 364, 2019
|
|
6J5K
 
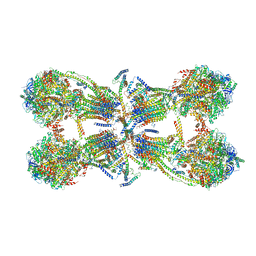 | | Cryo-EM structure of the mammalian ATP synthase tetramer bound with inhibitory protein IF1 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP synthase F1 subunit alpha, ... | | Authors: | Gu, J, Zhang, L, Yi, J, Yang, M. | | Deposit date: | 2019-01-11 | | Release date: | 2019-06-26 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (6.2 Å) | | Cite: | Cryo-EM structure of the mammalian ATP synthase tetramer bound with inhibitory protein IF1.
Science, 364, 2019
|
|
6J5J
 
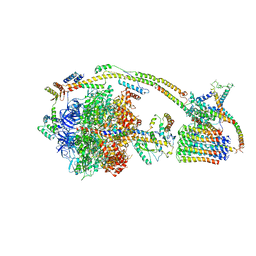 | | Cryo-EM structure of the mammalian E-state ATP synthase | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP synthase F1 subunit epsilon, ... | | Authors: | Gu, J, Zhang, L, Yi, J, Yang, M. | | Deposit date: | 2019-01-11 | | Release date: | 2019-06-26 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.45 Å) | | Cite: | Cryo-EM structure of the mammalian ATP synthase tetramer bound with inhibitory protein IF1.
Science, 364, 2019
|
|
7Y8A
 
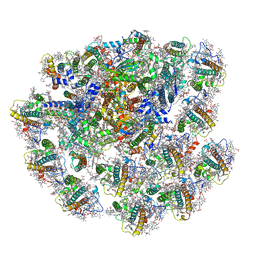 | | Cryo-EM structure of cryptophyte photosystem I | | Descriptor: | (1~{R})-3,5,5-trimethyl-4-[(3~{E},5~{E},7~{E},9~{E},11~{E},13~{E},15~{E})-3,7,12,16-tetramethyl-18-[(4~{R})-2,6,6-trimethyl-4-oxidanyl-cyclohexen-1-yl]octadeca-3,5,7,9,11,13,15-heptaen-1,17-diynyl]cyclohex-3-en-1-ol, (1~{R})-3,5,5-trimethyl-4-[(3~{E},5~{E},7~{E},9~{E},11~{E},13~{E},15~{E},17~{E})-3,7,12,16-tetramethyl-18-(2,6,6-trimethylcyclohexen-1-yl)octadeca-3,5,7,9,11,13,15,17-octaen-1-ynyl]cyclohex-3-en-1-ol, (1~{R})-3,5,5-trimethyl-4-[(3~{E},5~{E},7~{E},9~{E},11~{E},13~{E},15~{E},17~{E})-3,7,12,16-tetramethyl-18-[(1~{R},4~{R})-2,6,6-trimethyl-4-oxidanyl-cyclohex-2-en-1-yl]octadeca-3,5,7,9,11,13,15,17-octaen-1-ynyl]cyclohex-3-en-1-ol, ... | | Authors: | Zhao, L.S, Zhang, Y.Z, Liu, L.N, Li, K. | | Deposit date: | 2022-06-23 | | Release date: | 2023-04-12 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (2.71 Å) | | Cite: | Structural basis and evolution of the photosystem I-light-harvesting supercomplex of cryptophyte algae.
Plant Cell, 35, 2023
|
|
7Y7B
 
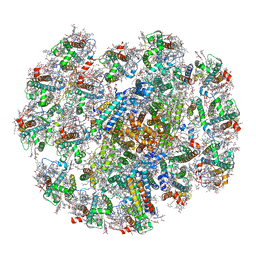 | | Cryo-EM structure of cryptophyte photosystem I | | Descriptor: | (1~{R})-3,5,5-trimethyl-4-[(3~{E},5~{E},7~{E},9~{E},11~{E},13~{E},15~{E})-3,7,12,16-tetramethyl-18-[(4~{R})-2,6,6-trimethyl-4-oxidanyl-cyclohexen-1-yl]octadeca-3,5,7,9,11,13,15-heptaen-1,17-diynyl]cyclohex-3-en-1-ol, (1~{R})-3,5,5-trimethyl-4-[(3~{E},5~{E},7~{E},9~{E},11~{E},13~{E},15~{E},17~{E})-3,7,12,16-tetramethyl-18-(2,6,6-trimethylcyclohexen-1-yl)octadeca-3,5,7,9,11,13,15,17-octaen-1-ynyl]cyclohex-3-en-1-ol, (1~{R})-3,5,5-trimethyl-4-[(3~{E},5~{E},7~{E},9~{E},11~{E},13~{E},15~{E},17~{E})-3,7,12,16-tetramethyl-18-[(1~{R},4~{R})-2,6,6-trimethyl-4-oxidanyl-cyclohex-2-en-1-yl]octadeca-3,5,7,9,11,13,15,17-octaen-1-ynyl]cyclohex-3-en-1-ol, ... | | Authors: | Zhao, L.S, Li, K, Zhang, Y.Z, Liu, L.N. | | Deposit date: | 2022-06-22 | | Release date: | 2023-04-12 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (2.66 Å) | | Cite: | Structural basis and evolution of the photosystem I-light-harvesting supercomplex of cryptophyte algae.
Plant Cell, 35, 2023
|
|
1K2N
 
 | |
8IK0
 
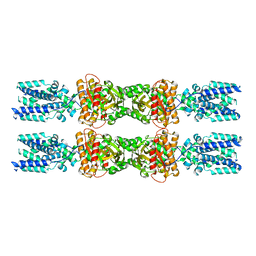 | |
8IK3
 
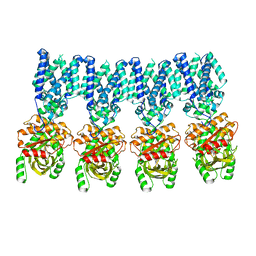 | |
7YR5
 
 | |
