8FHF
 
 | |
8FKE
 
 | |
6J02
 
 | | Crystal structure of the SRCR domain of mouse SCARA1 | | Descriptor: | CALCIUM ION, Macrophage scavenger receptor types I and II | | Authors: | Cheng, C, Hu, Z. | | Deposit date: | 2018-12-20 | | Release date: | 2019-11-13 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.803 Å) | | Cite: | The scavenger receptor SCARA1 (CD204) recognizes dead cells through spectrin.
J.Biol.Chem., 294, 2019
|
|
6J9A
 
 | | Crystal structure of Arabidopsis thaliana VAL1 in complex with FLC DNA fragment | | Descriptor: | B3 domain-containing transcription repressor VAL1, DNA (5'-D(*AP*AP*TP*CP*CP*AP*TP*GP*CP*AP*GP*AP*AP*TP*C)-3'), DNA (5'-D(*AP*TP*TP*CP*TP*GP*CP*AP*TP*GP*GP*AP*TP*TP*G)-3') | | Authors: | Hu, H, Du, J. | | Deposit date: | 2019-01-22 | | Release date: | 2019-05-29 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.915 Å) | | Cite: | Embryonic resetting of the parental vernalized state by two B3 domain transcription factors in Arabidopsis.
Nat.Plants, 5, 2019
|
|
6J9C
 
 | | Crystal structure of Arabidopsis thaliana transcription factor LEC2-DNA complex | | Descriptor: | B3 domain-containing transcription factor LEC2, DNA (5'-D(*CP*AP*AP*TP*CP*CP*AP*TP*GP*CP*AP*GP*AP*AP*T)-3'), DNA (5'-D(*GP*AP*TP*TP*CP*TP*GP*CP*AP*TP*GP*GP*AP*TP*T)-3') | | Authors: | Hu, H, Du, J. | | Deposit date: | 2019-01-22 | | Release date: | 2019-05-29 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.102 Å) | | Cite: | Embryonic resetting of the parental vernalized state by two B3 domain transcription factors in Arabidopsis.
Nat.Plants, 5, 2019
|
|
6J94
 
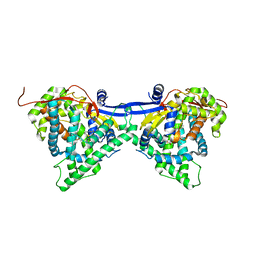 | | Crystal structure of CYP97A3 | | Descriptor: | PROTOPORPHYRIN IX CONTAINING FE, Protein LUTEIN DEFICIENT 5, chloroplastic | | Authors: | Niu, G, Guo, Q, Wang, J, Zhao, S. | | Deposit date: | 2019-01-22 | | Release date: | 2020-01-22 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.401 Å) | | Cite: | Structural basis for plant lutein biosynthesis from alpha-carotene.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6J95
 
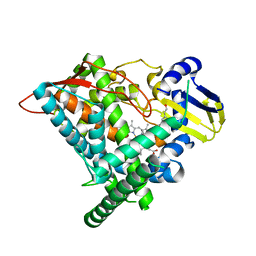 | | Crystal structure of CYP97A3 in complex with retinal | | Descriptor: | PROTOPORPHYRIN IX CONTAINING FE, Protein LUTEIN DEFICIENT 5, chloroplastic, ... | | Authors: | Niu, G, Guo, Q, Wang, J, Zhao, S. | | Deposit date: | 2019-01-22 | | Release date: | 2020-01-22 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.002 Å) | | Cite: | Structural basis for plant lutein biosynthesis from alpha-carotene.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6JGF
 
 | |
6JGV
 
 | |
6JGW
 
 | |
6JNI
 
 | |
8HB9
 
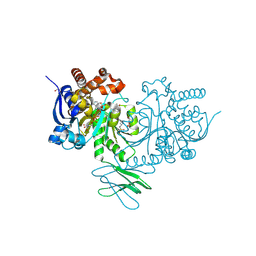 | | Crystal Structure of Human IDH1 R132H Mutant in Complex with NADPH and Compound IHMT-IDH1-053 | | Descriptor: | DI(HYDROXYETHYL)ETHER, GLYCEROL, Isocitrate dehydrogenase [NADP] cytoplasmic, ... | | Authors: | Guo, G, Wang, B, Liu, J, Liu, Q. | | Deposit date: | 2022-10-27 | | Release date: | 2023-05-24 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure-based discovery of IHMT-IDH1-053 as a potent irreversible IDH1 mutant selective inhibitor.
Eur.J.Med.Chem., 256, 2023
|
|
6J9B
 
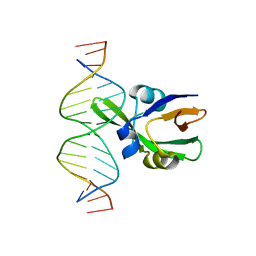 | | Arabidopsis FUS3-DNA complex | | Descriptor: | B3 domain-containing transcription factor FUS3, DNA (5'-D(*AP*AP*TP*CP*CP*AP*TP*GP*CP*AP*GP*AP*AP*TP*C)-3'), DNA (5'-D(*AP*TP*TP*CP*TP*GP*CP*AP*TP*GP*GP*AP*TP*TP*G)-3') | | Authors: | Hu, H, Du, J. | | Deposit date: | 2019-01-22 | | Release date: | 2019-05-29 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Embryonic resetting of the parental vernalized state by two B3 domain transcription factors in Arabidopsis.
Nat.Plants, 5, 2019
|
|
6JGX
 
 | | Crystal structure of the transcriptional regulator CadR from P. putida in complex with Cadmium(II) and DNA | | Descriptor: | CADMIUM ION, CadR, DNA (5'-D(*CP*AP*CP*CP*CP*TP*AP*TP*AP*GP*TP*GP*GP*CP*TP*AP*CP*AP*GP*GP*GP*T)-3'), ... | | Authors: | Liu, X.C, Gan, J.H, Chen, H. | | Deposit date: | 2019-02-15 | | Release date: | 2019-09-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.71 Å) | | Cite: | Selective cadmium regulation mediated by a cooperative binding mechanism in CadR.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6L8I
 
 | | Crystal structure of CYP97A3 mutant S290D/W300L/S304V | | Descriptor: | PROTOPORPHYRIN IX CONTAINING FE, Protein LUTEIN DEFICIENT 5, chloroplastic | | Authors: | Niu, G, Guo, Q, Liu, L. | | Deposit date: | 2019-11-06 | | Release date: | 2020-06-10 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural basis for plant lutein biosynthesis from alpha-carotene.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
7W09
 
 | | UGT74AN2, Plant Steroid Glycosyltransferase | | Descriptor: | Glycosyltransferase | | Authors: | Wei, H, Feng, L. | | Deposit date: | 2021-11-18 | | Release date: | 2022-03-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Functional and Structural Dissection of a Plant Steroid 3-O-Glycosyltransferase Facilitated the Engineering Enhancement of Sugar Donor Promiscuity
Acs Catalysis, 2022
|
|
7W11
 
 | | glycosyltransferase | | Descriptor: | DIGITOXIGENIN, Glycosyltransferase, URIDINE-5'-DIPHOSPHATE, ... | | Authors: | Wei, H, Feng, L. | | Deposit date: | 2021-11-19 | | Release date: | 2022-03-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Functional and Structural Dissection of a Plant Steroid 3-O-Glycosyltransferase Facilitated the Engineering Enhancement of Sugar Donor Promiscuity
Acs Catalysis, 2022
|
|
7W0Z
 
 | | Glycosyltranferase UGT74AN2 | | Descriptor: | 5-[(1R,2S,4R,6R,7R,10S,11S,14S,16R)-14-hydroxy-7,11-dimethyl-3-oxapentacyclo[8.8.0.02,4.02,7.011,16]octadecan-6-yl]pyran-2-one, Glycosyltransferase, URIDINE-5'-DIPHOSPHATE | | Authors: | Wei, H, Feng, L. | | Deposit date: | 2021-11-18 | | Release date: | 2022-03-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Functional and Structural Dissection of a Plant Steroid 3-O-Glycosyltransferase Facilitated the Engineering Enhancement of Sugar Donor Promiscuity
Acs Catalysis, 2022
|
|
7W1H
 
 | | glycosyltransferase | | Descriptor: | Glycosyltransferase, URIDINE-5'-DIPHOSPHATE-GLUCOSE | | Authors: | Wei, H, Feng, L. | | Deposit date: | 2021-11-19 | | Release date: | 2022-03-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Functional and Structural Dissection of a Plant Steroid 3-O-Glycosyltransferase Facilitated the Engineering Enhancement of Sugar Donor Promiscuity
Acs Catalysis, 2022
|
|
7W10
 
 | | UGT74AN2 | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Glycosyltransferase, URIDINE-5'-DIPHOSPHATE, ... | | Authors: | Wei, H, Long, F. | | Deposit date: | 2021-11-18 | | Release date: | 2022-03-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Functional and Structural Dissection of a Plant Steroid 3-O-Glycosyltransferase Facilitated the Engineering Enhancement of Sugar Donor Promiscuity
Acs Catalysis, 2022
|
|
7W0K
 
 | | plant glycosyltransferase | | Descriptor: | GLYCEROL, Glycosyltransferase, URIDINE-5'-DIPHOSPHATE | | Authors: | Wei, H, Feng, L. | | Deposit date: | 2021-11-18 | | Release date: | 2022-03-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Functional and Structural Dissection of a Plant Steroid 3-O-Glycosyltransferase Facilitated the Engineering Enhancement of Sugar Donor Promiscuity
Acs Catalysis, 2022
|
|
7W1B
 
 | | Glycosyltransferase | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, DIGITOXIGENIN, Glycosyltransferase, ... | | Authors: | Wei, H, Feng, L. | | Deposit date: | 2021-11-19 | | Release date: | 2022-03-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Functional and Structural Dissection of a Plant Steroid 3-O-Glycosyltransferase Facilitated the Engineering Enhancement of Sugar Donor Promiscuity
Acs Catalysis, 2022
|
|
8ZOJ
 
 | |
7ET5
 
 | | Crystal structure of Arabidopsis TEM1 AP2 domain | | Descriptor: | AP2/ERF and B3 domain-containing transcription repressor TEM1, SULFATE ION | | Authors: | Hu, H, Du, J. | | Deposit date: | 2021-05-12 | | Release date: | 2021-09-15 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.052 Å) | | Cite: | TEM1 combinatorially binds to FLOWERING LOCUS T and recruits a Polycomb factor to repress the floral transition in Arabidopsis.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7ET6
 
 | | Crystal structure of Arabidopsis TEM1 B3-DNA complex | | Descriptor: | AP2/ERF and B3 domain-containing transcription repressor TEM1, FT-RY14-F, FT-RY14-R | | Authors: | Hu, H, Du, J. | | Deposit date: | 2021-05-12 | | Release date: | 2021-09-15 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | TEM1 combinatorially binds to FLOWERING LOCUS T and recruits a Polycomb factor to repress the floral transition in Arabidopsis.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
