8WLL
 
 | | Cryo-EM structure of human VMAT2 Y422C, in the presence of reserpine, determined in an inward-facing conformation | | Descriptor: | Synaptic vesicular amine transporter, reserpine | | Authors: | Qu, Q, Wang, Y, Zhou, Z. | | Deposit date: | 2023-09-30 | | Release date: | 2024-06-12 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.74 Å) | | Cite: | Transport and inhibition mechanism for VMAT2-mediated synaptic vesicle loading of monoamines.
Cell Res., 34, 2024
|
|
8WLK
 
 | | Cryo-EM structure of human VMAT2 in presence of Tetrabenazine, determined in an outward-facing conformation | | Descriptor: | (3S,5R,11bS)-9,10-dimethoxy-3-(2-methylpropyl)-1,3,4,6,7,11b-hexahydro-2H-pyrido[2,1-a]isoquinolin-2-one, Synaptic vesicular amine transporter | | Authors: | Qu, Q, Wang, Y, Zhou, Z. | | Deposit date: | 2023-09-30 | | Release date: | 2024-06-12 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.37 Å) | | Cite: | Transport and inhibition mechanism for VMAT2-mediated synaptic vesicle loading of monoamines.
Cell Res., 34, 2024
|
|
8WLM
 
 | |
8WM5
 
 | |
5X5Y
 
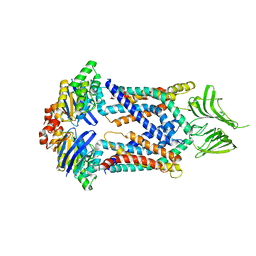 | | A membrane protein complex | | Descriptor: | Probable ATP-binding component of ABC transporter, Uncharacterized protein | | Authors: | Luo, Q, Yang, X, Huang, Y. | | Deposit date: | 2017-02-18 | | Release date: | 2017-04-05 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.465 Å) | | Cite: | Structural basis for lipopolysaccharide extraction by ABC transporter LptB2FG
Nat. Struct. Mol. Biol., 24, 2017
|
|
4B61
 
 | | In meso structure of alginate transporter, AlgE, from Pseudomoas aeruginosa, PAO1. Crystal form 3. | | Descriptor: | (2R)-2,3-DIHYDROXYPROPYL(7Z)-PENTADEC-7-ENOATE, (2S)-2,3-DIHYDROXYPROPYL(7Z)-PENTADEC-7-ENOATE, ACETATE ION, ... | | Authors: | Tan, J, Pye, V.E, Aragao, D, Caffrey, M. | | Deposit date: | 2012-08-08 | | Release date: | 2013-07-17 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.402 Å) | | Cite: | A Conformational Landscape for Alginate Secretion Across the Outer Membrane of Pseudomonas Aeruginosa.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4AZL
 
 | | In meso structure of alginate transporter, AlgE, from Pseudomoas aeruginosa, PAO1, crystal form 2. | | Descriptor: | (2R)-2,3-DIHYDROXYPROPYL(7Z)-PENTADEC-7-ENOATE, (2S)-2,3-DIHYDROXYPROPYL(7Z)-PENTADEC-7-ENOATE, ALGINATE PRODUCTION PROTEIN ALGE, ... | | Authors: | Tan, J, Pye, V.E, Aragao, D, Caffrey, M. | | Deposit date: | 2012-06-26 | | Release date: | 2013-07-03 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | A Conformational Landscape for Alginate Secretion Across the Outer Membrane of Pseudomonas Aeruginosa.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
8Y16
 
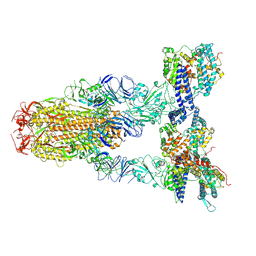 | | Cryo-EM structure of SARS-CoV-2 Omicron JN.1 spike protein in complex with human ACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, ... | | Authors: | Li, L.J, Gu, Y.H, Qi, J.X, Gao, G.F. | | Deposit date: | 2024-01-23 | | Release date: | 2024-07-03 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.98 Å) | | Cite: | Spike structures, receptor binding, and immune escape of recently circulating SARS-CoV-2 Omicron BA.2.86, JN.1, EG.5, EG.5.1, and HV.1 sub-variants.
Structure, 32, 2024
|
|
8XLV
 
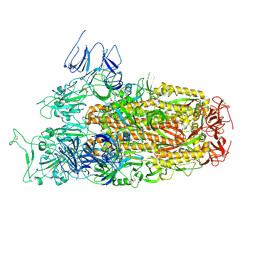 | | Cryo-EM structure of SARS-CoV-2 Omicron BA.2.86 spike protein(6P), 1-RBD-up state | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Li, L.J, Gu, Y.H, Shi, K.Y, Qi, J.X, Gao, G.F. | | Deposit date: | 2023-12-26 | | Release date: | 2024-07-03 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (3.07 Å) | | Cite: | Spike structures, receptor binding, and immune escape of recently circulating SARS-CoV-2 Omicron BA.2.86, JN.1, EG.5, EG.5.1, and HV.1 sub-variants.
Structure, 32, 2024
|
|
8Y5J
 
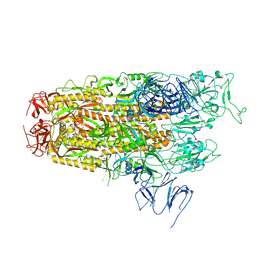 | | Cryo-EM structure of SARS-CoV-2 Omicron JN.1 spike protein | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Li, L.J, Gu, Y.H, Qi, J.X, Gao, G.F. | | Deposit date: | 2024-01-31 | | Release date: | 2024-07-03 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (2.94 Å) | | Cite: | Spike structures, receptor binding, and immune escape of recently circulating SARS-CoV-2 Omicron BA.2.86, JN.1, EG.5, EG.5.1, and HV.1 sub-variants.
Structure, 32, 2024
|
|
8I2N
 
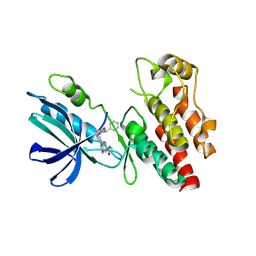 | | The RIPK1 kinase domain in complex with QY7-2B compound | | Descriptor: | Receptor-interacting serine/threonine-protein kinase 1, ~{N}-methyl-1-[4-[[[1-methyl-5-(phenylmethyl)pyrazol-3-yl]carbonylamino]methyl]phenyl]benzimidazole-5-carboxamide | | Authors: | Gong, X.Y, Li, Y, Meng, H.Y, Pan, L.F. | | Deposit date: | 2023-01-14 | | Release date: | 2024-01-31 | | Last modified: | 2024-08-21 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Structure-based development of potent and selective type-II kinase inhibitors of RIPK1.
Acta Pharm Sin B, 14, 2024
|
|
4LX4
 
 | | Crystal Structure Determination of Pseudomonas stutzeri endoglucanase Cel5A using a Twinned Data Set | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Endoglucanase(Endo-1,4-beta-glucanase)protein | | Authors: | Dutoit, R, Delsaute, M, Berlemont, R, Van Elder, D, Galleni, M, Bauvois, C. | | Deposit date: | 2013-07-29 | | Release date: | 2014-07-30 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.556 Å) | | Cite: | Crystal structure determination of Pseudomonas stutzeri A1501 endoglucanase Cel5A: the search for a molecular basis for glycosynthesis in GH5_5 enzymes.
Acta Crystallogr D Struct Biol, 75, 2019
|
|
8XWD
 
 | | Croy-EM structure of alpha synuclein fibril with EGCG | | Descriptor: | (2R,3R)-5,7-dihydroxy-2-(3,4,5-trihydroxyphenyl)-3,4-dihydro-2H-chromen-3-yl 3,4,5-trihydroxybenzoate, Alpha-synuclein | | Authors: | Li, X, Liu, C. | | Deposit date: | 2024-01-16 | | Release date: | 2025-02-26 | | Last modified: | 2025-04-30 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Single-Molecule Insight Into alpha-Synuclein Fibril Structure and Mechanics Modulated by Chemical Compounds.
Adv Sci, 12, 2025
|
|
7OTJ
 
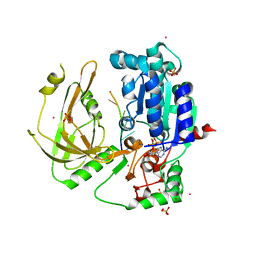 | | Crystal structure of Pif1 helicase from Candida albicans | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATP-dependent DNA helicase PIF1, DNA (5'-D(P*TP*TP*TP*TP*TP*T)-3'), ... | | Authors: | Rety, S, Xi, X.G. | | Deposit date: | 2021-06-10 | | Release date: | 2021-07-07 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.58 Å) | | Cite: | Structural study of the function of Candida Albicans Pif1.
Biochem.Biophys.Res.Commun., 567, 2021
|
|
3U28
 
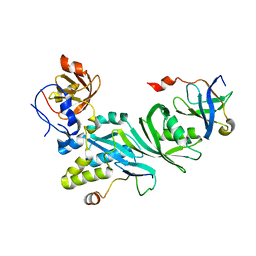 | | Crystal structure of a Cbf5-Nop10-Gar1 complex from Saccharomyces cerevisiae | | Descriptor: | H/ACA ribonucleoprotein complex subunit 1, H/ACA ribonucleoprotein complex subunit 3, H/ACA ribonucleoprotein complex subunit 4 | | Authors: | Ye, K, Li, S. | | Deposit date: | 2011-10-02 | | Release date: | 2011-12-14 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Reconstitution and structural analysis of the yeast box H/ACA RNA-guided pseudouridine synthase
Genes Dev., 25, 2011
|
|
7TEI
 
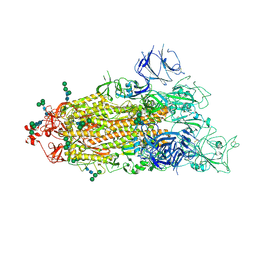 | |
7TF8
 
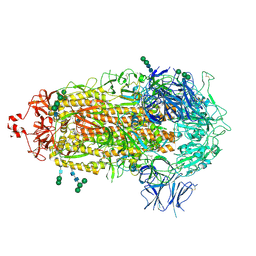 | |
7TL9
 
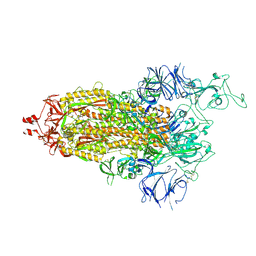 | |
7THT
 
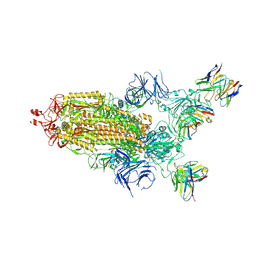 | | CryoEM structure of SARS-CoV-2 S protein in complex with Receptor Binding Domain antibody DH1042 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, DH1042 heavy chain, ... | | Authors: | Manne, K, May, A, Acharya, P. | | Deposit date: | 2022-01-12 | | Release date: | 2022-02-16 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.42 Å) | | Cite: | Structural diversity of the SARS-CoV-2 Omicron spike.
Mol.Cell, 82, 2022
|
|
7TL1
 
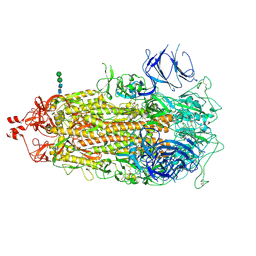 | |
5O6D
 
 | | Structure of ScPif1 in complex with polydT and ATPgS | | Descriptor: | ATP-dependent DNA helicase PIF1, DNA (5'-D(P*TP*TP*TP*TP*TP*T)-3'), MAGNESIUM ION, ... | | Authors: | Lu, K.Y, Chen, W.F, Rety, S, Liu, N.N, Xu, X.G. | | Deposit date: | 2017-06-06 | | Release date: | 2017-12-13 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.283 Å) | | Cite: | Insights into the structural and mechanistic basis of multifunctional S. cerevisiae Pif1p helicase.
Nucleic Acids Res., 46, 2018
|
|
6AHF
 
 | | CryoEM Reconstruction of Hsp104 N728A Hexamer | | Descriptor: | Heat shock protein 104, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER | | Authors: | Zhang, X, Zhang, L, Zhang, S. | | Deposit date: | 2018-08-17 | | Release date: | 2019-02-13 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (6.78 Å) | | Cite: | Heat shock protein 104 (HSP104) chaperones soluble Tau via a mechanism distinct from its disaggregase activity.
J. Biol. Chem., 294, 2019
|
|
5O6B
 
 | | Structure of ScPif1 in complex with GGGTTTT and ADP-AlF4 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATP-dependent DNA helicase PIF1, DNA (5'-D(*GP*GP*GP*TP*TP*T)-3'), ... | | Authors: | Lu, K.Y, Chen, W.F, Rety, S, Liu, N.N, Xu, X.G. | | Deposit date: | 2017-06-06 | | Release date: | 2017-12-13 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.029 Å) | | Cite: | Insights into the structural and mechanistic basis of multifunctional S. cerevisiae Pif1p helicase.
Nucleic Acids Res., 46, 2018
|
|
5O6E
 
 | | Structure of ScPif1 in complex with TTTGGGTT and ADP-AlF4 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATP-dependent DNA helicase PIF1, CHLORIDE ION, ... | | Authors: | Lu, K.Y, Chen, W.F, Rety, S, Liu, N.N, Xu, X.G. | | Deposit date: | 2017-06-06 | | Release date: | 2017-12-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.345 Å) | | Cite: | Insights into the structural and mechanistic basis of multifunctional S. cerevisiae Pif1p helicase.
Nucleic Acids Res., 46, 2018
|
|
6LLB
 
 | | Crystal structure of mpy-RNase J (mutant S247A), an archaeal RNase J from Methanolobus psychrophilus R15, in complex with 6 nt RNA | | Descriptor: | MPY-RNase J, RNA (5'-R(P*AP*AP*AP*AP*AP*A)-3'), SULFATE ION, ... | | Authors: | Li, D.F, Hou, Y.J, Guo, L. | | Deposit date: | 2019-12-22 | | Release date: | 2020-01-01 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | A newly identified duplex RNA unwinding activity of archaeal RNase J depends on processive exoribonucleolysis coupled steric occlusion by its structural archaeal loops.
Rna Biol., 17, 2020
|
|
