5VOX
 
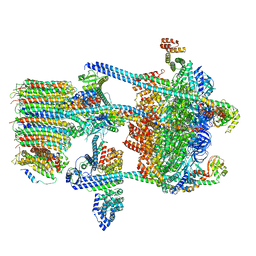 | | Yeast V-ATPase in complex with Legionella pneumophila effector SidK (rotational state 1) | | Descriptor: | V-type proton ATPase catalytic subunit A,V-type proton ATPase catalytic subunit A, V-type proton ATPase subunit B, V-type proton ATPase subunit C, ... | | Authors: | Zhao, J. | | Deposit date: | 2017-05-03 | | Release date: | 2017-06-21 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (6.8 Å) | | Cite: | Molecular basis for the binding and modulation of V-ATPase by a bacterial effector protein.
PLoS Pathog., 13, 2017
|
|
4GZG
 
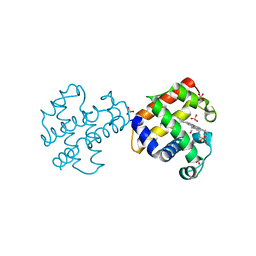 | | Crystal structures of DHPA-CO complex | | Descriptor: | CARBON MONOXIDE, Dehaloperoxidase A, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Zhao, J, Franzen, S. | | Deposit date: | 2012-09-06 | | Release date: | 2013-09-11 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | The role of distal histidine in carbonmonoxide DHP structure
To be Published
|
|
3J9T
 
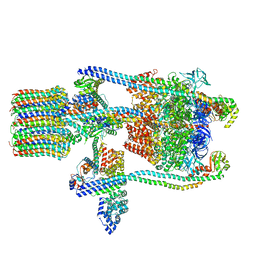 | | Yeast V-ATPase state 1 | | Descriptor: | V-type proton ATPase catalytic subunit A, V-type proton ATPase subunit B, V-type proton ATPase subunit C, ... | | Authors: | Zhao, J, Benlekbir, S, Rubinstein, J.L. | | Deposit date: | 2015-02-23 | | Release date: | 2015-05-13 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (6.9 Å) | | Cite: | Electron cryomicroscopy observation of rotational states in a eukaryotic V-ATPase.
Nature, 521, 2015
|
|
3J9V
 
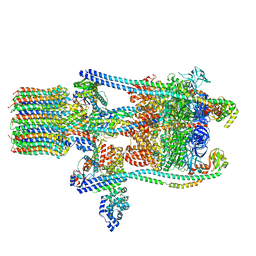 | | Yeast V-ATPase state 3 | | Descriptor: | V-type proton ATPase catalytic subunit A, V-type proton ATPase subunit B, V-type proton ATPase subunit C, ... | | Authors: | Zhao, J, Benlekbir, S, Rubinstein, J.L. | | Deposit date: | 2015-02-23 | | Release date: | 2015-05-13 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (8.3 Å) | | Cite: | Electron cryomicroscopy observation of rotational states in a eukaryotic V-ATPase.
Nature, 521, 2015
|
|
7YAC
 
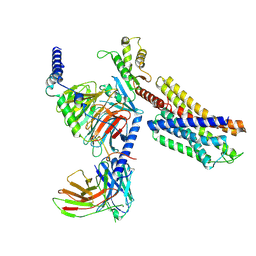 | | Paltusotine-bound SSTR2-Gi complex | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(i) subunit alpha-1, ... | | Authors: | Zhao, J, Shao, Z. | | Deposit date: | 2022-06-27 | | Release date: | 2023-04-19 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.24 Å) | | Cite: | Prospect of acromegaly therapy: molecular mechanism of clinical drugs octreotide and paltusotine.
Nat Commun, 14, 2023
|
|
7YAE
 
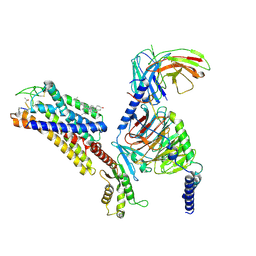 | | Octreotide-bound SSTR2-Gi complex | | Descriptor: | CHOLESTEROL, DPN-CYS-PHE-DTR-LYS-THR-CYS-THO, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Zhao, J, Shao, Z. | | Deposit date: | 2022-06-28 | | Release date: | 2023-04-19 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.37 Å) | | Cite: | Prospect of acromegaly therapy: molecular mechanism of clinical drugs octreotide and paltusotine.
Nat Commun, 14, 2023
|
|
8V02
 
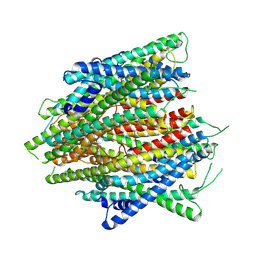 | | AaegOR10 structure bound to o-cresol | | Descriptor: | Odorant receptor OR10, Odorant receptor Orco, o-cresol | | Authors: | Zhao, J, del Marmol, J. | | Deposit date: | 2023-11-16 | | Release date: | 2024-06-19 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural basis of odor sensing by insect heteromeric odorant receptors.
Science, 384, 2024
|
|
8V00
 
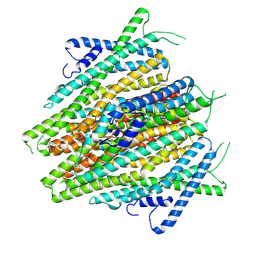 | | AaegOR10 apo structure | | Descriptor: | Odorant receptor OR10, Odorant receptor Orco | | Authors: | Zhao, J, del Marmol, J. | | Deposit date: | 2023-11-16 | | Release date: | 2024-06-19 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (2.88 Å) | | Cite: | Structural basis of odor sensing by insect heteromeric odorant receptors.
Science, 384, 2024
|
|
8V3D
 
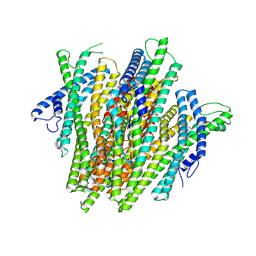 | | AgamOR28 structure bound to 2,4,5-trimethylthiazole | | Descriptor: | 2,4,5-trimethyl-1,3-thiazole, OR28, Odorant receptor Orco | | Authors: | Zhao, J, del Marmol, J. | | Deposit date: | 2023-11-27 | | Release date: | 2024-06-19 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (2.95 Å) | | Cite: | Structural basis of odor sensing by insect heteromeric odorant receptors.
Science, 384, 2024
|
|
8V3C
 
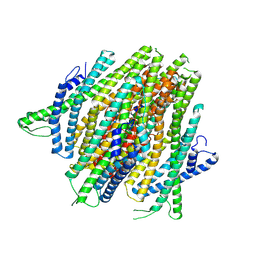 | | AgamOR28 structure without ligand | | Descriptor: | OR28, Odorant receptor Orco | | Authors: | Zhao, J, del Marmol, J. | | Deposit date: | 2023-11-27 | | Release date: | 2024-06-19 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (2.62 Å) | | Cite: | Structural basis of odor sensing by insect heteromeric odorant receptors.
Science, 384, 2024
|
|
9GBW
 
 | | Overall structure of the substrate-bound U11 snRNP | | Descriptor: | P120-5'SS, Programmed cell death protein 7, Small nuclear ribonucleoprotein E, ... | | Authors: | Zhao, J, Galej, W.P. | | Deposit date: | 2024-07-31 | | Release date: | 2025-02-05 | | Last modified: | 2025-02-19 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structural basis of 5' splice site recognition by the minor spliceosome.
Mol.Cell, 85, 2025
|
|
9GC0
 
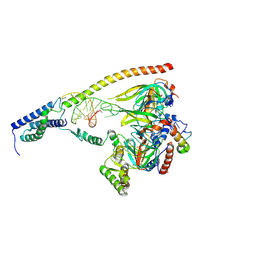 | | 3'-lobe of the substrate-bound U11 snRNP | | Descriptor: | Programmed cell death protein 7, Small nuclear ribonucleoprotein E, Small nuclear ribonucleoprotein F, ... | | Authors: | Zhao, J, Galej, W.P. | | Deposit date: | 2024-07-31 | | Release date: | 2025-02-05 | | Last modified: | 2025-02-19 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural basis of 5' splice site recognition by the minor spliceosome.
Mol.Cell, 85, 2025
|
|
9GCM
 
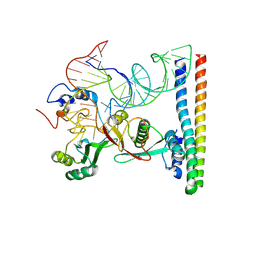 | | Structure of the U11 snRNP core | | Descriptor: | Programmed cell death protein 7, U11 snRNA, U11/U12 small nuclear ribonucleoprotein 25 kDa protein, ... | | Authors: | Zhao, J, Galej, W.P. | | Deposit date: | 2024-08-02 | | Release date: | 2025-02-05 | | Last modified: | 2025-02-19 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural basis of 5' splice site recognition by the minor spliceosome.
Mol.Cell, 85, 2025
|
|
9GBZ
 
 | | 5'-lobe of the substrate-bound U11 snRNP | | Descriptor: | P120-5'SS, U11 snRNA, U11/U12 small nuclear ribonucleoprotein 25 kDa protein, ... | | Authors: | Zhao, J, Galej, W.P. | | Deposit date: | 2024-07-31 | | Release date: | 2025-02-05 | | Last modified: | 2025-02-19 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural basis of 5' splice site recognition by the minor spliceosome.
Mol.Cell, 85, 2025
|
|
9GCL
 
 | | Structure of the U11 snRNP C-lobe | | Descriptor: | Small nuclear ribonucleoprotein E, Small nuclear ribonucleoprotein F, Small nuclear ribonucleoprotein G, ... | | Authors: | Zhao, J, Galej, W.P. | | Deposit date: | 2024-08-02 | | Release date: | 2025-02-05 | | Last modified: | 2025-02-19 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural basis of 5' splice site recognition by the minor spliceosome.
Mol.Cell, 85, 2025
|
|
4JYQ
 
 | | DHP-CO crystal structure | | Descriptor: | CARBON MONOXIDE, Dehaloperoxidase A, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Zhao, J, Srajer, V, Franzen, S. | | Deposit date: | 2013-03-31 | | Release date: | 2014-04-23 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Observation of protein valves for the control of the internal flow of small molecules using time-resolved X-ray crystallography
To be Published
|
|
8G4G
 
 | | Crystal Engineering with One 8-mer DNA | | Descriptor: | DNA (5'-D(*AP*TP*CP*GP*G)-3'), DNA (5'-D(*AP*TP*CP*GP*GP*CP*CP*G)-3'), DNA (5'-D(P*CP*CP*G)-3') | | Authors: | Zhao, J, Zhang, C, Lu, B, Seeman, N.C, Noinaj, N, Sha, R, Mao, C. | | Deposit date: | 2023-02-09 | | Release date: | 2023-05-10 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Divergence and Convergence: Complexity Emerges in Crystal Engineering from an 8-mer DNA.
J.Am.Chem.Soc., 145, 2023
|
|
7CFP
 
 | |
7CFQ
 
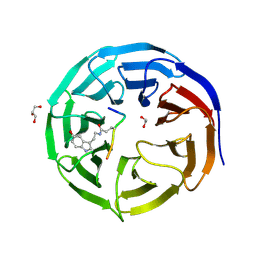 | | Crystal structure of WDR5 in complex with H3K4me3Q5ser peptide | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, H3K4me3Q5ser peptide, ... | | Authors: | Zhao, J, Zhang, X, Zang, J. | | Deposit date: | 2020-06-27 | | Release date: | 2021-07-07 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural insights into the recognition of histone H3Q5 serotonylation by WDR5.
Sci Adv, 7, 2021
|
|
8Y56
 
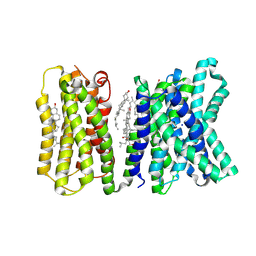 | | Cryo-EM reveals cholesterol binding in the lysosomal GPCR-like protein LYCHOS | | Descriptor: | CHOLESTEROL, Lysosomal cholesterol signaling protein, SODIUM ION, ... | | Authors: | Zhao, J, Shen, Q.Y, Zhang, Y, Shao, Z.H. | | Deposit date: | 2024-01-31 | | Release date: | 2025-01-29 | | Last modified: | 2025-06-04 | | Method: | ELECTRON MICROSCOPY (2.83 Å) | | Cite: | Cryo-EM reveals cholesterol binding in the lysosomal GPCR-like protein LYCHOS.
Nat.Struct.Mol.Biol., 32, 2025
|
|
5KF4
 
 | |
5J2Y
 
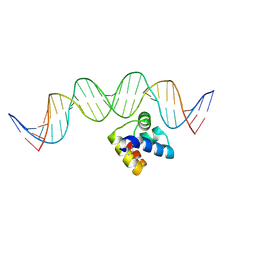 | | Molecular insight into the regulatory mechanism of the quorum-sensing repressor RsaL in Pseudomonas aeruginosa | | Descriptor: | DNA (26-MER), Regulatory protein | | Authors: | Zhao, J, Gan, J, Zhang, J, Kang, H, Kong, W, Zhu, M, Li, F, Song, Y, Qin, J, Liang, H. | | Deposit date: | 2016-03-30 | | Release date: | 2017-04-12 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of Pseudomonas aeruginosa RsaL bound to promoter DNA reaffirms its role as a global regulator involved in quorum-sensing.
Nucleic Acids Res., 45, 2017
|
|
6ZT9
 
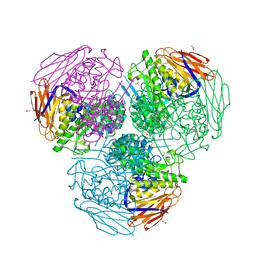 | | X-ray structure of mutated arabinofuranosidase | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ACETATE ION, ... | | Authors: | Tandrup, T, Lo Leggio, L, Zhao, J, Bissaro, B, Barbe, S, Andre, I, Dumon, C, O'Donohue, M.J, Faure, R. | | Deposit date: | 2020-07-17 | | Release date: | 2021-02-10 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Probing the determinants of the transglycosylation/hydrolysis partition in a retaining alpha-l-arabinofuranosidase.
N Biotechnol, 62, 2021
|
|
6ZTA
 
 | | X-ray structure of mutated arabinofuranosidase | | Descriptor: | Alpha-L-arabinofuranosidase | | Authors: | Tandrup, T, Lo Leggio, L, Zhao, J, Bissaro, B, Barbe, S, Andre, I, Dumon, C, O'Donohue, M.J, Faure, R. | | Deposit date: | 2020-07-17 | | Release date: | 2021-02-10 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Probing the determinants of the transglycosylation/hydrolysis partition in a retaining alpha-l-arabinofuranosidase.
N Biotechnol, 62, 2021
|
|
6ZT8
 
 | | X-ray structure of mutated arabinofuranosidase | | Descriptor: | Alpha-L-arabinofuranosidase, CHLORIDE ION, PENTAETHYLENE GLYCOL, ... | | Authors: | Tandrup, T, Lo Leggio, L, Zhao, J, Bissaro, B, Barbe, S, Andre, I, Dumon, C, O'Donohue, M.J, Faure, R. | | Deposit date: | 2020-07-17 | | Release date: | 2021-02-10 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Probing the determinants of the transglycosylation/hydrolysis partition in a retaining alpha-l-arabinofuranosidase.
N Biotechnol, 62, 2021
|
|
